Viruses have all kinds of tricks up their sleeves. It often does not take long for pathogens such as SARS-CoV-2 to adapt and escape immune recognition. Viruses with mutations, that evade immune recognition more efficiently than the original virus will have a selective advantage and prevail over time, increasingly dominating the infection landscape. “If the new virus variant can successfully escape the immune response, it is no longer sufficient to have recovered from one of the previous variants or to have been vaccinated with a previously effective vaccine,” says Prof. Luka Cicin-Sain, head of the department “Viral Immunology” at the HZI. “With vaccine development, we are always playing catch-up with the spread of new escape variants. That's the very nature of virus evolution. Nevertheless, we need to outsmart the viruses and reduce their advantages, both for currently circulating ones and for those that will be around in future pandemics.”
A swift identification of mutations that are crucial for the immune escape enable the rapid adaptation of vaccines to new virus variants. In their current study, the team led by Cicin-Sain presents a promising new approach for doing just that. It is based on a previously established method called mutational scanning: Each mutation found in the new variant is introduced separately into the original virus to generate a library of virus mutants. The effects of each individual mutation can thus be defined by comparing the mutants from the library to the original and to the new virus variant. However, in their study, the HZI researchers modified the procedure by introducing the mutations one by one into the new virus variant to make it resemble the original virus at each position. Hence, they used the methodology in reverse, which is why they called it reverse mutational scanning.
How reverse mutational scanning works
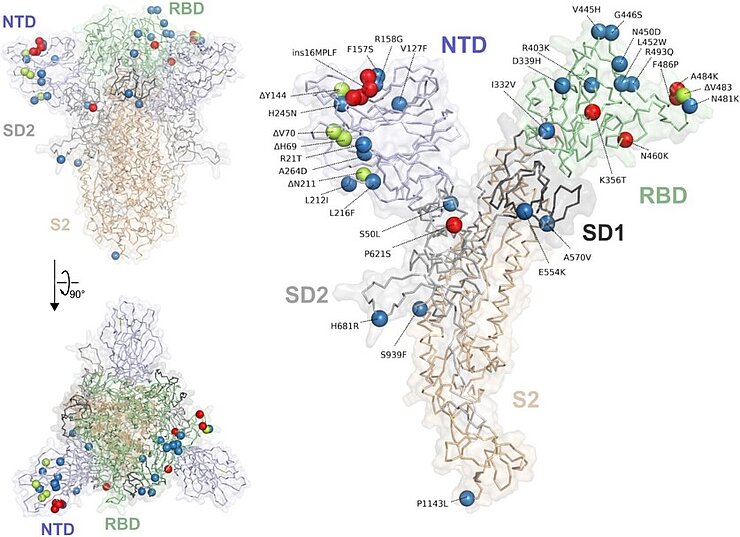
What exactly is the process behind reverse mutational scanning and how did the research team go about it? To test their new approach, the scientists examined viral variants of SARS-CoV-2 as an example. They wanted to find out which of the 33 mutations that distinguish the BA.2.86 virus variant from the original BA.2 variant are responsible for immune escape.