Antibiotic resistance is rising worldwide and poses a major challenge to healthcare systems. Understanding the underlying processes of how and why some bacteria develop such defenses is critical. Differences in bacterial gene expression—in which the information in a gene is read—could provide clues. Bacterial single-cell RNA sequencing, or “scRNA-seq”, has become an essential tool for analyzing these variations. This technology provides a detailed picture of gene expression in a single cell at a specific point in time. However, the method generates huge amounts of data that are difficult for researchers to visualize and analyze. Accordingly, there is a great need for new approaches and tools to display and understand this information.
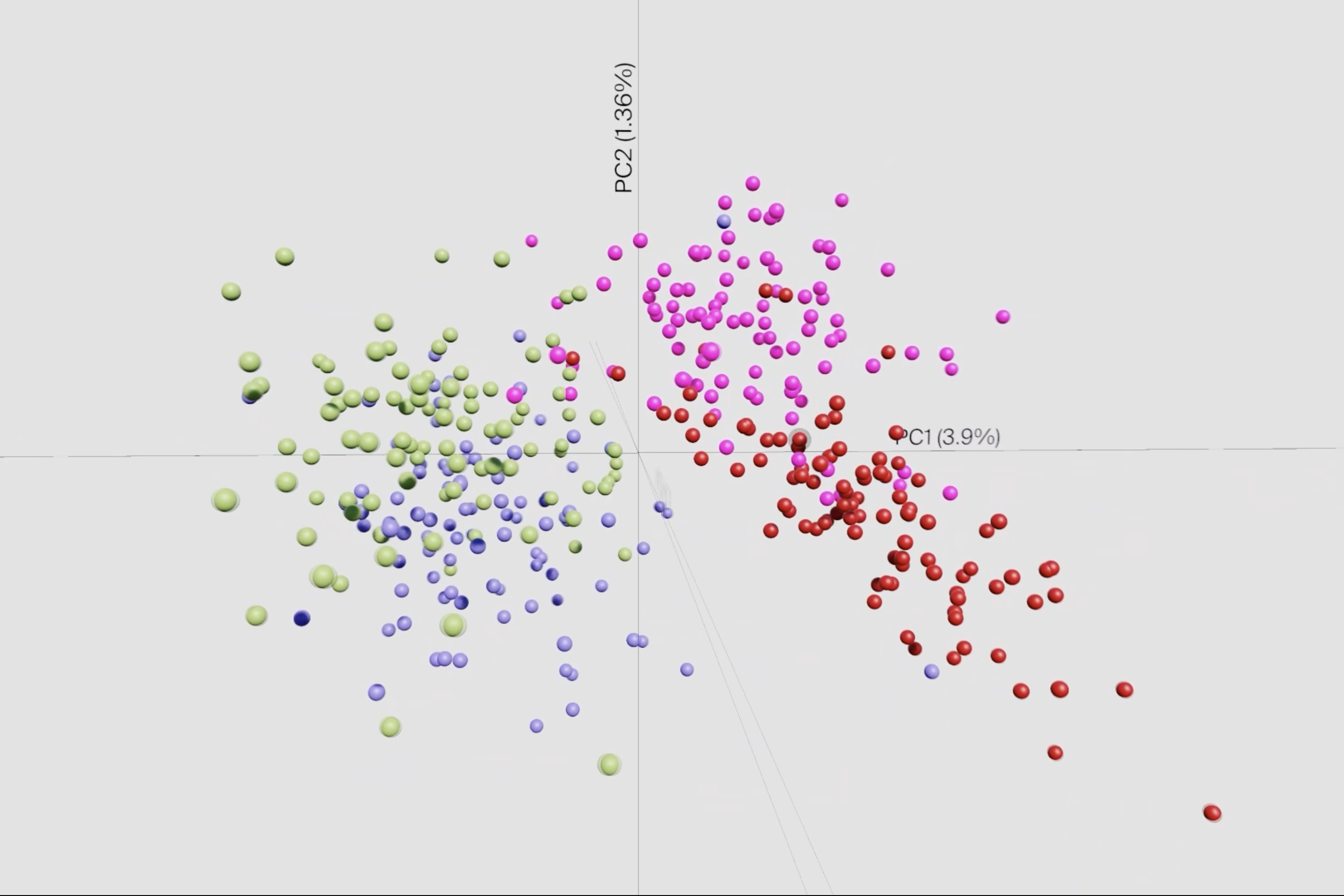
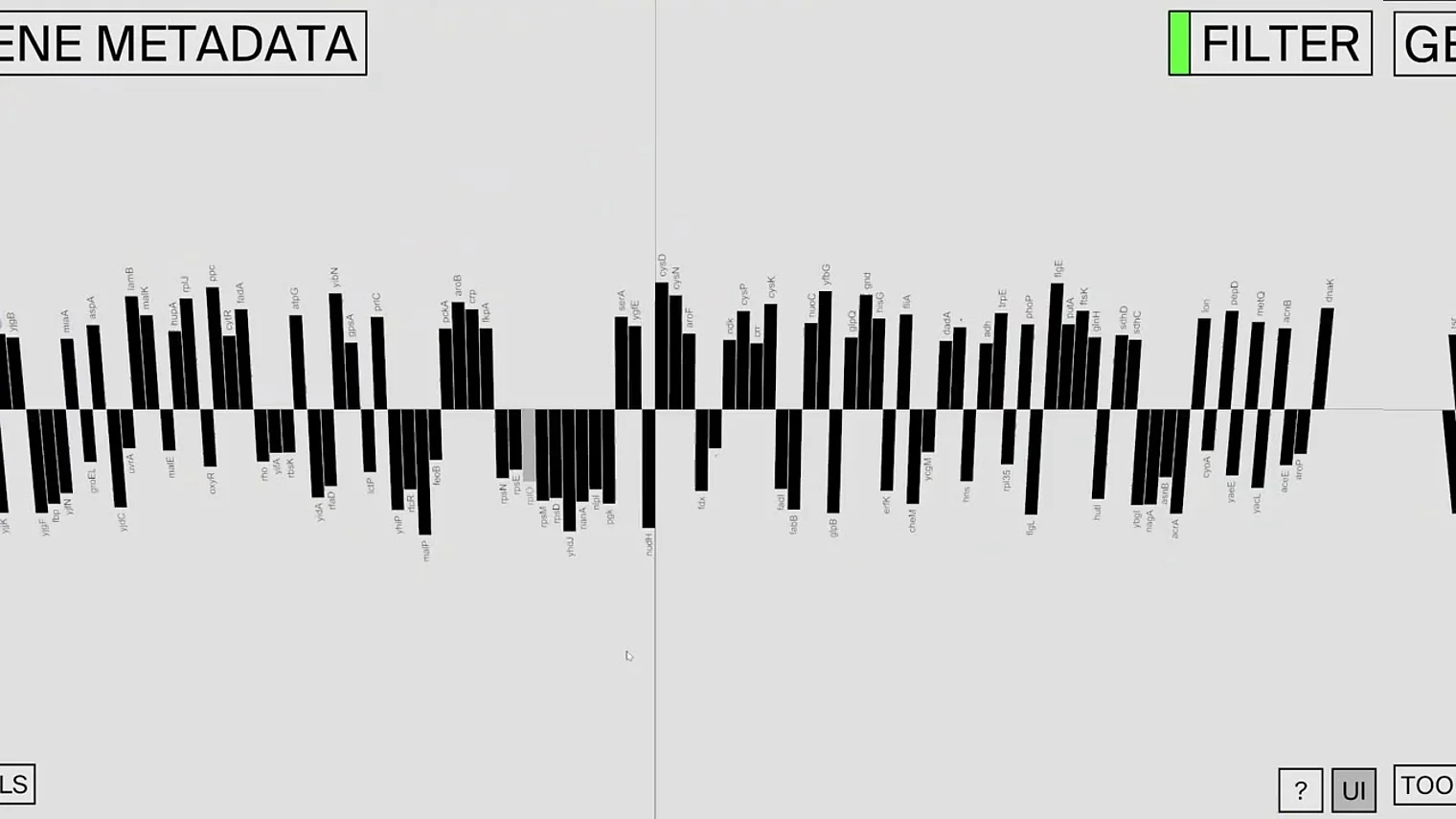
Researchers at the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg, a site of the Helmholtz Centre for Infection Research (HZI) in Braunschweig in cooperation with the Julius-Maximilians-Universität Würzburg (JMU), and designers at the Technical University of Applied Sciences Würzburg-Schweinfurt (THWS) have now developed such a tool. Their desktop application, called sCIRCLE (single-Cell Interactive Real-time Computer visualization for Low-dimensional Exploration), enables interactive 3D visualization of scRNA-seq data. It allows users to view single cells, enriched with metadata, from different angles and in real-time.