Anti-infectives from Microbiota

Our research
The spread of antibiotic-resistant human pathogenic microorganisms poses an increasing threat to human health, and the development of new anti-infective agents and a better understanding of their function and mode of action is urgently needed.
The department MICA pursues a microbiota-based strategy to identify new active substances from microorganisms.
What does "microbiota" mean?
Microbial communities (microbiota) are composed of a large number of different bacteria, fungi and representatives of unicellular and few-celled eukaryotes, as well as viruses.
Microbiota are found, among other things, on human, animal and plant tissue surfaces, where they can assume essential functions for the host. In many cases, the composition of the microbiota correlates with its localization and thus its function.
What role do natural substances play?
Microorganisms regulate and manipulate their coexistence by emitting bioactive natural substances. Microbial natural products can act as antibiotics to protect producers, but can also act as a cellular signal, act as a morphogen for the host organism, or being metabolized as a nutrient. However, the chemical structures of many of these modulating natural substances are unknown, and their natural function, as well as their influence on the microbiota and possible application potentials are only very little known to date.
How can we harvest the biosynthetic potential of micoorganisms?
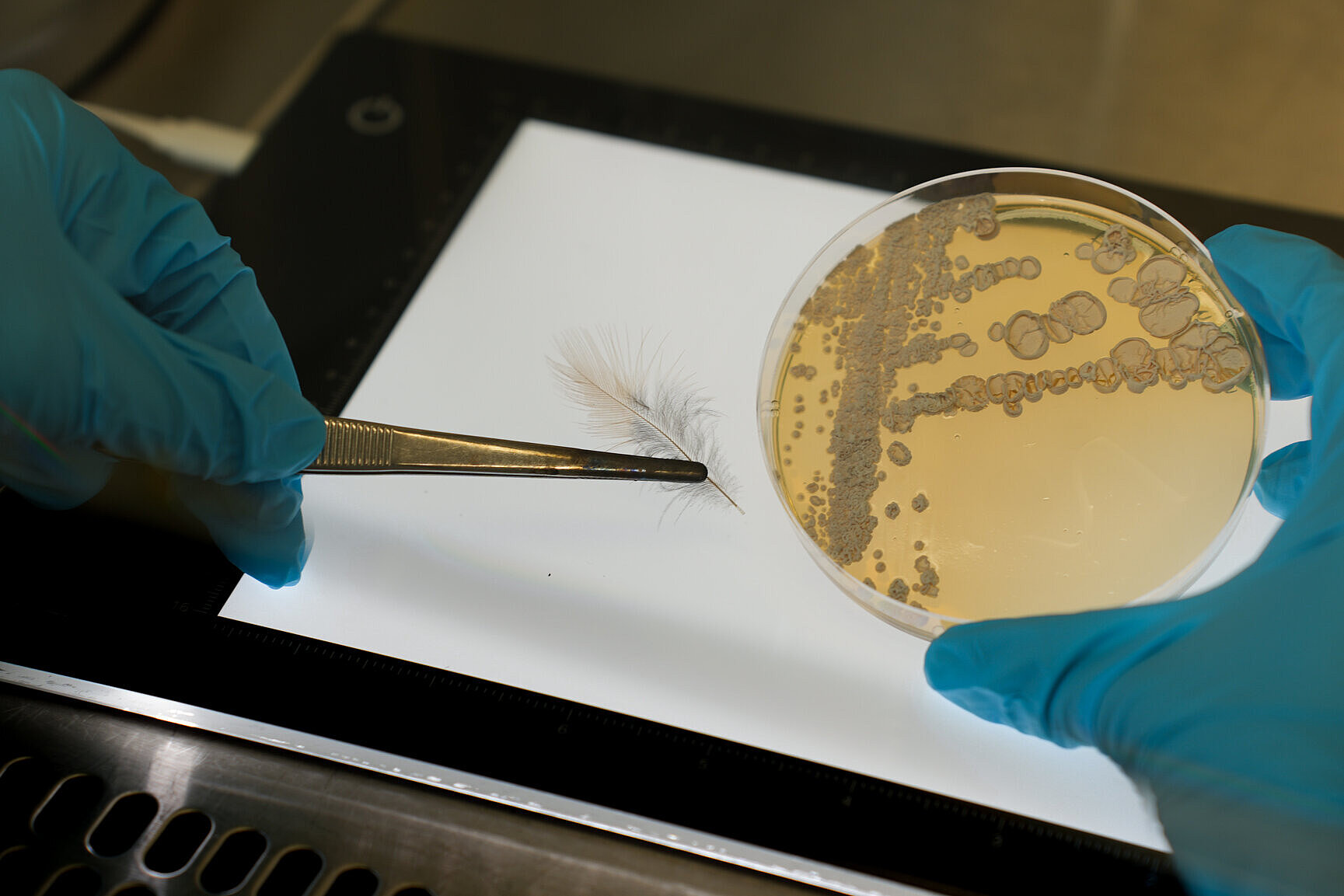
Since natural products play important roles in microbial interactions, their production is closely linked to the composition of the microbiota. The Beemelmanns group analyzes representative microbial communities to unlock this chemical space.
Target-oriented bioassays based on bacteria-bacteria and bacteria-fungus interaction studies are being used to stimulate the production of bioactive natural products. The group uses established and innovative analytical methods to elucidate the structure of the secreted natural substances.
The resulting natural products are synthesized for their anti-infective mode of action and natural products with high potency to better understand their structure-activity relationships and advance the profiling of the most promising candidates.
In addition, the influence of the characterized natural substances on symbiotic communities is investigated using microbiome studies. With the help of these studies, we hope to be able to make better statements about the stability and dynamics of microbial communities.
The elucidation of the biosynthetic pathways of new natural products and their regulation is also an important part of our research in order to improve our biocatalytic understanding and advance the development of biotechnological approaches.
Members of the department have different scientific educational backgrounds, which includes analytical and synthetic chemistry as well as microbiology, biochemistry and pharmacy.
Our research
The spread of antibiotic-resistant human pathogenic microorganisms poses an increasing threat to human health, and the development of new anti-infective agents and a better understanding of their function and mode of action is urgently needed.
The department MICA pursues a microbiota-based strategy to identify new active substances from microorganisms.
What does "microbiota" mean?
Microbial communities (microbiota) are composed of a large number of different bacteria, fungi and representatives of unicellular and few-celled eukaryotes, as well as viruses.
Microbiota are found, among other things, on human, animal and plant tissue surfaces, where they can assume essential functions for the host. In many cases, the composition of the microbiota correlates with its localization and thus its function.
What role do natural substances play?
Microorganisms regulate and manipulate their coexistence by emitting bioactive natural substances. Microbial natural products can act as antibiotics to protect producers, but can also act as a cellular signal, act as a morphogen for the host organism, or being metabolized as a nutrient. However, the chemical structures of many of these modulating natural substances are unknown, and their natural function, as well as their influence on the microbiota and possible application potentials are only very little known to date.
How can we harvest the biosynthetic potential of micoorganisms?
Since natural products play important roles in microbial interactions, their production is closely linked to the composition of the microbiota. The Beemelmanns group analyzes representative microbial communities to unlock this chemical space.
Target-oriented bioassays based on bacteria-bacteria and bacteria-fungus interaction studies are being used to stimulate the production of bioactive natural products. The group uses established and innovative analytical methods to elucidate the structure of the secreted natural substances.
The resulting natural products are synthesized for their anti-infective mode of action and natural products with high potency to better understand their structure-activity relationships and advance the profiling of the most promising candidates.
In addition, the influence of the characterized natural substances on symbiotic communities is investigated using microbiome studies. With the help of these studies, we hope to be able to make better statements about the stability and dynamics of microbial communities.
The elucidation of the biosynthetic pathways of new natural products and their regulation is also an important part of our research in order to improve our biocatalytic understanding and advance the development of biotechnological approaches.
Members of the department have different scientific educational backgrounds, which includes analytical and synthetic chemistry as well as microbiology, biochemistry and pharmacy.
Prof Christine Beemelmanns
We use microbial interaction studies, molecular biology and synthetic approaches to advance the identification and production of drug candidates from natural products.

Christine Beemelmanns studied Chemistry at the RWTH Aachen, and after graduation went to Japan for a one-year research stay in the group of Prof. Sodeoka at RIKEN. Back in Germany she worked at the FU Berlin with Prof. Reißig and received her PhD in Organic Chemistry. She then worked another six month in Japan at the University of Tokyo under the supervision of Prof K. Suzuki and joined shortly afterwards the group of Prof. Clardy at Harvard Medical School (Boston) in 2011.
End of 2013, she received a call from the Hans-Knöll Institute (HKI) to work there as a Junior Research Group leader in the field of Natural Products Chemistry and Chemical Biology.
In 2020, she was elected as Margaret L. and Harlan L. Goering Visiting Professors in Organic Chemistry at UW Madison (spring term 2020).
In 2022 was appointed Professor for Biochemistry of Microbial Metabolism at the Leipzig University. In October 2022 she accepted a call for the Full Professorship on Medicinal-Pharmaceutical Microbiota Research at Saarland University together with the HIPS.
Her research combines different aspects of natural product chemistry, applied microbiology and organic and natural product chemistry and aims to chemically and to functionally characterize microbial signaling and defense molecules in different model systems. The analysis of ancient and evolved microbial interactions allows her to discover unprecedented chemical core structures with potential pharmaceutical potential.
Selected Publications
Raguž L,# Peng CC,# Rutaganira FUN, Krüger T, Stanisic A, Jautzus T, Kries H, Kniemeyer O, Brakhage A, King N, Beemelmanns C* (2022) Total synthesis and functional evaluation of IORs, sulfonolipid-based inhibitors of cell differentiation in Salpingoeca rosetta. Angew Chem Int Ed, e202209105.
Raguž L, Peng CC, Kaiser M, Görls H, Beemelmanns C* (2022) A modular approach to the antifungal Sphingofungin family: Concise total synthesis of Sphingofungin A and C. Angew Chem Int Ed, e202112616
Guo H, Rischer M, Westermann M, Beemelmanns C* (2021) Two distinct bacterial biofilm components trigger metamorphosis in the colonial hydrozoan Hydractinia echinata. mBio 12(3), e0040121.
Schalk F, Gostinčar C, Kreuzenbeck NB, Conlon BH, Sommerwerk E, Rabe P, Burkhardt I, Krüger T, Kniemeyer O, Brakhage AA, Gunde-Cimerman N, de Beer ZW, Dickschat JS, Poulsen M, Beemelmanns C* (2021) The termite fungal cultivar Termitomyces combines diverse enzymes and oxidative reactions for plant biomass conversion. mBio 12(3), e0355120.
Guo H, Schwitalla JW, Benndorf R, Baunach M, Steinbeck C, Görls H, de Beer ZW, Regestein L, Beemelmanns C* (2020) Gene cluster activation in a bacterial symbiont leads to halogenated angucyclic maduralactomycins and spirocyclic actinospirols. Org Lett 22(7), 2634-2638
A complete list of publications can be found on the HIPS website
![[Translate to English:] Bakterienkolonien](/fileadmin/_processed_/8/f/csm_co_culture_a4550343c1.jpg)