Antiviral and Antivirulence Drugs

Our Research
We progress our antiinfective projects through all the stages of drug discovery. Our hit-finding strategies are selected individually for the target molecule at hand. In the case of receptors and enzymes with pockets druggable with small molecules (e.g. PqsR, PqsE), we employ fragment-based methodologies and transform identified starting points (hits) into drug-like molecules (leads) by medicinal chemistry optimization. In this regard, we follow a multi-parameter optimization considering not only potency but also pharmacokinetic and safety pharmacology endpoints. Ideally, leads are optimized towards preclinical profiling candidate status in order to facilitate preclinical development.
Difficult-to-drug macromolecule-macromolecule interactions provide challenging, yet, promising and notoriously underexploited points of disease intervention. We have been successful in applying above-mentioned fragment-based hit identification to a protein-DNA interaction essential for the lifecycle of human herpesvirus 8 (Kaposi´s sarcoma herpesvirus, KSHV) mediated by the latency-associated nuclear antigen (LANA). In order to supplement and complement these small molecule-directed approaches, we also apply the phage display technology for the screening of very short macrocyclic (constrained) peptide sequences. The resulting hits are readily synthetically accessible starting points amenable to medicinal chemistry optimization towards novel drug molecules at the boarders of Lipinski´s rule of five (or just slightly beyond). In the future, we aim to combine fragment- and phage-display-based approaches to generate peptide-small molecules hybrids with the aim to explore currently untapped chemical space.
Our Research
We progress our antiinfective projects through all the stages of drug discovery. Our hit-finding strategies are selected individually for the target molecule at hand. In the case of receptors and enzymes with pockets druggable with small molecules (e.g. PqsR, PqsE), we employ fragment-based methodologies and transform identified starting points (hits) into drug-like molecules (leads) by medicinal chemistry optimization. In this regard, we follow a multi-parameter optimization considering not only potency but also pharmacokinetic and safety pharmacology endpoints. Ideally, leads are optimized towards preclinical profiling candidate status in order to facilitate preclinical development.
Difficult-to-drug macromolecule-macromolecule interactions provide challenging, yet, promising and notoriously underexploited points of disease intervention. We have been successful in applying above-mentioned fragment-based hit identification to a protein-DNA interaction essential for the lifecycle of human herpesvirus 8 (Kaposi´s sarcoma herpesvirus, KSHV) mediated by the latency-associated nuclear antigen (LANA). In order to supplement and complement these small molecule-directed approaches, we also apply the phage display technology for the screening of very short macrocyclic (constrained) peptide sequences. The resulting hits are readily synthetically accessible starting points amenable to medicinal chemistry optimization towards novel drug molecules at the boarders of Lipinski´s rule of five (or just slightly beyond). In the future, we aim to combine fragment- and phage-display-based approaches to generate peptide-small molecules hybrids with the aim to explore currently untapped chemical space.
Prof Dr Martin Empting
As academic drug discovery scientists we have a clear mandate to drive innovation in anti-infective research. We achieve this is by unlocking and exploiting new modes of action for the treatment of viral and bacterial infections through medicinal chemistry methods.

Dr. Martin Empting studied chemistry at the Technical University Darmstadt. After receiving his Diploma, which was awarded with the Prize of the “Dr. Anton-Keller-Stiftung”, he performed his PhD studies under supervision of Prof. Dr. Harald Kolmar. In 2013, he graduated with summa cum laude and his doctoral thesis focusing on peptidic drug candidates and biomimetic concepts was awarded with the Prize of the "Familie Bottling-Stiftung". After a PostDoc with Prof. Dr. Rolf W. Hartmann at the Helmholtz Institute for Pharmaceutical Research Saarland (HIPS) where he focused on the design and optimization of novel anti-infective agents against Gram-negative bacteria, he now conducts his own independent research. His central aim is to devise novel synthetic drug molecules for the development of urgently needed antiinfective therapies.
Selected Publications
A. Berwanger, S. C. Stein, A. M. Kany, M. Gartner, B. Loretz, C.-M. Lehr, A. K. H. Hirsch, T. F. Schulz, M. Empting, Disrupting Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) latent replication with a small molecule inhibitor, Journal of Medicinal Chemistry (2023), 66, 10782-10790, https://doi.org/10.1021/acs.jmedchem.3c00990.
M. M. Hamed, A. S. Abdelsamie, K. Rox, C. Schütz, A. M. Kany, T. Röhrig, S. Schmelz, W. Blankenfeldt, A. Arce-Rodriguez, J. Manuel Borrero-de Acuña, D. Jahn, J. Rademacher, F. Ringshausen, B. Tümmler, A. K. H. Hirsch, R. W. Hartmann, M. Empting, Towards translation of PqsR inverse agonists: from in vitro efficacy optimization to in vivo proof-of-principle, Advanced Science (2023), 10, 2204443, https://doi.org/10.1002/advs.202204443.
V. Jakob, B. G. E. Zoller, J. Rinkes, Y. Wu, A. F. Kiefer, M. Hust, S. Polten, A. M. White, P. J. Harvey, T. Durek, D. J. Craik, A. Siebert, U. Kazmaier, M. Empting, Phage display-based discovery of cyclic peptides against the broad spectrum bacterial anti-virulence target CsrA, European Journal of Medicinal Chemistry (2022), 231, 114148, https://doi.org/10.1016/j.ejmech.2022.114148
C. Schütz, D.-K. Ho, M. Hamed, A. S. Abdelsamie, T. Röhrig, C. Herr, A. M. Kany, K. Rox, S. Schmelz, L. Siebenbürger, M. Wirth, C. Börger, S. Yahiaoui, R. Bals, A. Scrima, W. Blankenfeldt, J. C. Horstmann, R. Christmann, X. Murgia, M. Koch, A. Berwanger, B. Loretz, A. K. H. Hirsch, R. W. Hartmann, C.-M. Lehr, M. Empting, A new PqsR inverse agonist potentiates tobramycin efficacy to eradicate Pseudomonas aeruginosa biofilms, Advanced Science (2021), 8, 2004369, https://doi.org/10.1002/advs.202004369
P. Kirsch, V. Jakob, K. Oberhausen, S.C. Stein, I. Cucarro, T.F. Schulz, M. Empting, Fragment-based discovery of a qualified hit targeting the latency-associated nuclear antigen of the oncogenic Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8, Journal of Medicinal Chemistry (2019), 62, 3924-3939, https://doi.org/10.1021/acs.jmedchem.8b01827
A complete list of publications can be found on the HIPS website.
Technology Offers
The following technologies have been developed and patented by the department Antiviral and Antivirulence Drugs:
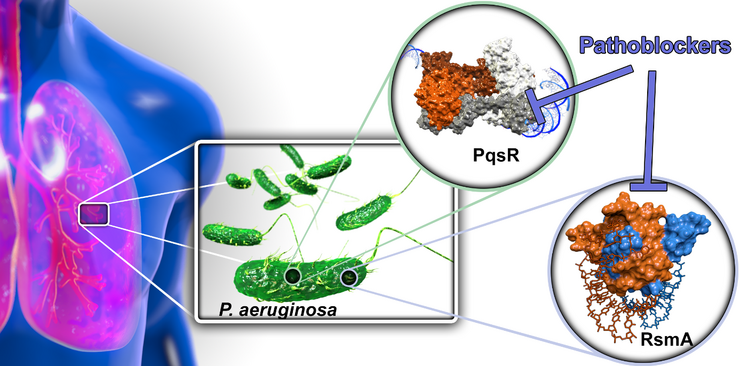
Targeting Bacterial Pathogenicity (Pathoblockers)
With the aim to devise novel treatment modalities and to divert from classical bactericidal or bacteriostatic antibiotics, we design and optimize synthetic molecules, which specifically block bacterial pathogenicity traits. In this regard, we address transcriptional and post-transcriptional regulators of bacterial virulence. By this means, we interfere with a multitude of virulence factors and do not interfere with bacterial viability. The overall aim of the project is to discover and advance compounds for the treatment of Pseudomonas aeruginosa infections, which is a problematic ESKAPE pathogen and the major cause of morbidity and mortality in patients with cystic fibrosis. To this end, we address to regulator systems: A) the Pseudomonas quinolone signal (PQS) quorum sensing (QS) system and B) the carbon storage regulator (Csr; also called regulator of secondary metabolites, Rsm) system.
A) QS provides a way for bacteria to communicate with each other via small diffusible signal molecules. This ability is essential for population-wide coordination of infection processes. We target the signal molecule receptor and transcriptional regulator PqsR (MvfR) by compounds acting as inverse agonists. The PQS QS system is quite unique resulting in a pathogen-specific mode-of-action, which preserves the commensal (beneficial) microbiota. By means, we successfully achieved lead status with three chemical classes of pathoblockers showing promising potency in the nanomolar range combined with a suitable pharmacokinetic and in vitro safety pharmacology profile. This project is currently in the late lead optimization stage.
B) In contrast to the PQS QS, the Csr/Rsm system is astonishingly widespread among pathogenic bacteria. Hence, this higher order regulatory system holds promise as a potential broad-spectrum pathoblocker target. We were able to identify identify natural product-derived small molecule-like hits as well as novel peptidic macrocyclic starting points for medicinal chemistry optimization. Demonstration of cellular activity mediated by blocking this post-transcriptional regulator is the next step in this project.
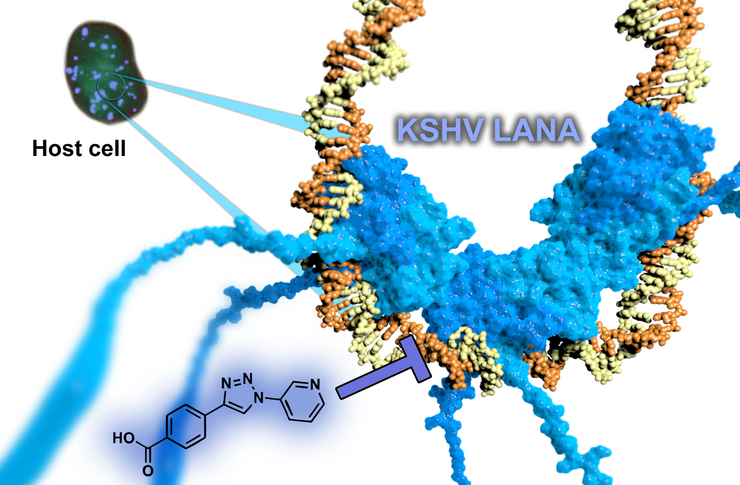
Targeting persistence of human Herpes Viruses
The latency-associated nuclear antigen (LANA) is required for latent replication and persistence of Kaposi’sarcoma-associated herpesvirus/human herpesvirus 8. LANA exerts important functions in the host cell like cell survival, transcriptional control, latent viral DNA replication, and stable episome segregation during mitosis. It acts via tethering the virus episome to the host chromatin. We aim to discover novel antiviral drugs, which to inhibit the interaction between LANA and the viral genome. By this means, we should be able to disrupt the latent replication of KSHV.
We were successful in finding the first LANA inhibitors by a fragment-based approach. The resulting optimized hit provides an ideal starting point for subsequent hit-to-lead campaigns providing evident target-binding, suitable ligand efficiencies, and favourable physicochemical properties. Further improved compounds are currently underway with a view to demonstrate antiviral activities in cell-based assays.