Bacterial Infection Ecology

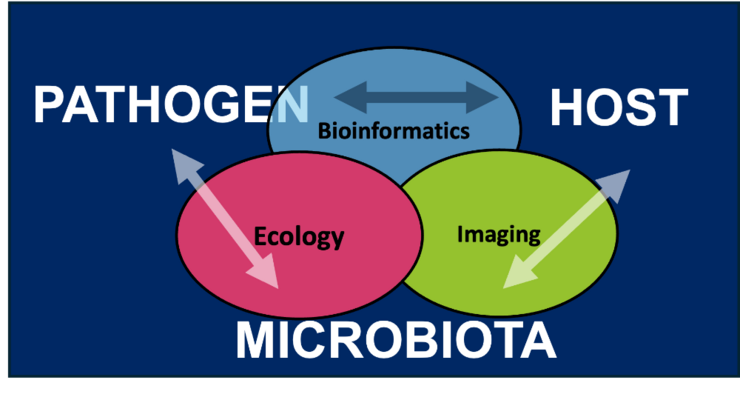
Our bodies are home to diverse microbial communities that protect us against pathogens, with the mucus-lined epithelia being a particularly dense area of colonisation. The Bacterial Infection Ecology Lab strives to identify the rules of engagement that govern the interaction between microbiota, pathogens and host cells within animals. Using a multidisciplinary approach that merges ecological theory, bioinformatics, and advanced imaging, our research focuses on three key themes:
Core Principles of Spatial Microbiome Organization
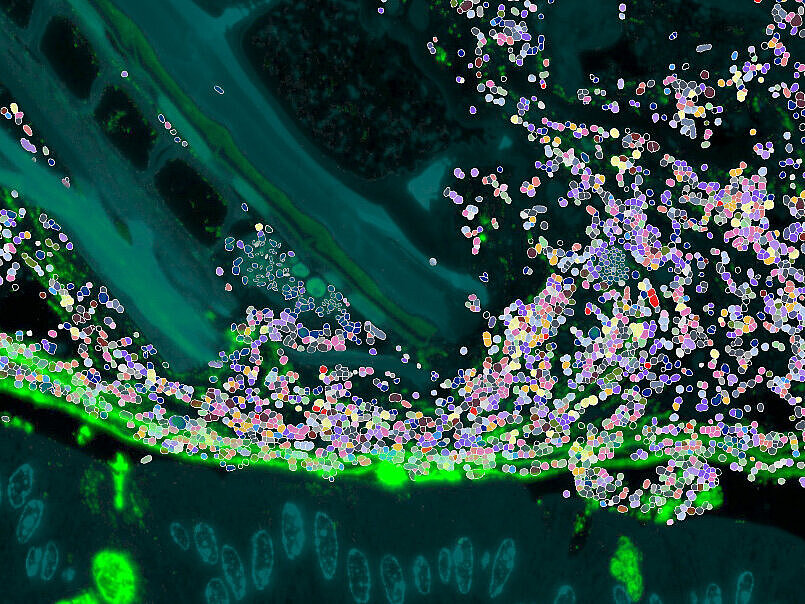
Microbial communities consist of many strains and species, and their spatial arrangement at the single-cell scale defines if they actually meet and interact. Current sequencing-based methods often fail to capture interactions in hosts at such fine scales, leaving us with a resolution gap where key ecological and evolutionary processes happen. Using new imaging technologies and defined spatial models, we are bridging this gap and resolve the spatial ecology of gut microbiomes at the cell level. Here, we systematically compare healthy and diseased states. These data will provide an important baseline on structural organisation principles of the gut microbiota and how they might be boosted to promote resilience to perturbations such as by antibiotics, inflammation and infection.
Mapping the impact of microbiota on infection outcomes

How do the microbiota prevent infection? In this line of research, we are investigating the spatial ecology of pathogen resistance, with a focus on gut infections and associated inflammation. To this end, we are mapping how microbiota-pathogen interactions alter infection outcomes. This will inform the identification of key microbiota for infection resistance and their underlying ecology in infection. It will also enable biogeography-guided reformulations of taxa and function to disentangle their precise role in infection. Ultimately, this hypothesis-driven research will provide new fundamental insights to guide the rational design of beneficial microbial communities to enhance infection resistance.
Host Manipulation & microbiome inspired therapeutics
How do gut microbiota manipulate their mammalian host? To this end, my lab screens the human gut microbiota using a combination of bioinformatics and cell assays. These insights into microbial community-mediated host manipulation have the potential to be of high translational value in identifying candidates for targeted immune therapies, as well as paves the way to symbiont-mediated immune modulation in the context of inflammation and infection.
Our bodies are home to diverse microbial communities that protect us against pathogens, with the mucus-lined epithelia being a particularly dense area of colonisation. The Bacterial Infection Ecology Lab strives to identify the rules of engagement that govern the interaction between microbiota, pathogens and host cells within animals. Using a multidisciplinary approach that merges ecological theory, bioinformatics, and advanced imaging, our research focuses on three key themes:
Core Principles of Spatial Microbiome Organization

Microbial communities consist of many strains and species, and their spatial arrangement at the single-cell scale defines if they actually meet and interact. Current sequencing-based methods often fail to capture interactions in hosts at such fine scales, leaving us with a resolution gap where key ecological and evolutionary processes happen. Using new imaging technologies and defined spatial models, we are bridging this gap and resolve the spatial ecology of gut microbiomes at the cell level. Here, we systematically compare healthy and diseased states. These data will provide an important baseline on structural organisation principles of the gut microbiota and how they might be boosted to promote resilience to perturbations such as by antibiotics, inflammation and infection.
Mapping the impact of microbiota on infection outcomes

How do the microbiota prevent infection? In this line of research, we are investigating the spatial ecology of pathogen resistance, with a focus on gut infections and associated inflammation. To this end, we are mapping how microbiota-pathogen interactions alter infection outcomes. This will inform the identification of key microbiota for infection resistance and their underlying ecology in infection. It will also enable biogeography-guided reformulations of taxa and function to disentangle their precise role in infection. Ultimately, this hypothesis-driven research will provide new fundamental insights to guide the rational design of beneficial microbial communities to enhance infection resistance.
Host Manipulation & microbiome inspired therapeutics
How do gut microbiota manipulate their mammalian host? To this end, my lab screens the human gut microbiota using a combination of bioinformatics and cell assays. These insights into microbial community-mediated host manipulation have the potential to be of high translational value in identifying candidates for targeted immune therapies, as well as paves the way to symbiont-mediated immune modulation in the context of inflammation and infection.
Our bodies are microbial ecosystems – resolving the interactions between us, our microbes and pathogens will guide innovative therapies to improve health and well-being.

Martin Jahn is a microbiologist, ecologist, and bioinformatician dedicated to understanding the intricate mechanisms of how microbes interact with our bodies and affect our health. He studied natural sciences with a focus on microbiology, ecology and bioinformatics at the University of Würzburg, Germany. During this time he conducted fieldwork on the spatial landscape ecology of Borrelia burgdorferi within host reservoirs as visiting scholar at Glasgow University with Prof Roman Biek.
Following his graduate studies, Martin Jahn was awarded a fellowship to pursue a Ph.D at the University of Kiel in the Collaborative Research Centre 1182 with Prof. Ute Hentschel. Here he studied the functional underpinnings of sponge symbioses, the oldest known animal-microbe symbiosis. His journey also took him to the University of Lausanne as a visiting scientist in 2018, where he worked with Professor Anders Meibom on correlative metabolic imaging.
In 2019, Martin Jahn was awarded his Ph.D. (summa cum laude), and in 2020 received a fellowship from the Human Frontier Science Programme (HFSP) to continue his scientific training at the University of Oxford. At Oxford, he worked as a postdoctoral researcher with Professor Kevin Foster, focusing on the biogeography of the gut microbiome and its implications for disease resistance. This involved establishing new technologies to map the cellular organisation of the entire host-associated microbiota and to study structural microbiome changes in different disease contexts. This experience further shaped his view of the human body as a complex and diverse ecosystem in which microbial interactions play a critical role in maintaining health.
Since October 2024, Martin Jahn is a group leader at the HZI Braunschweig, where he heads the Bacterial Infection Ecology group.
Team




Selected Publications
Jahn M.T., Arkhipova K. Markert S.M., Stigloher C., Lachnit T., Pita L., Kupczok A., Ribes M., Stengel S.T., Rosenstiel P., Dutilh B.E., Hentschel U. (2019). A Phage Protein Aids Bacterial Symbionts in Eukaryote Immune Evasion. Cell Host & Microbe; doi: 10.1016/j.chom.2019.08.019
Spragge F, Bakkeren E, Jahn, M.T., Araujo EBN, Pearson CF, Wang X, Pankhurst L, Cunrath O, Foster KR 2023 Microbiome diversity protects against pathogens by nutrient blocking. Science; doi: 10.1126/science.adj3502
Jahn, M.T., Lachnit, T., Markert, S.M., C. Stigloher, L. Pita, M. Ribes, B. E. Dutilh,. U. Hentschel (2021). Lifestyle of sponge symbiont phages by host prediction and correlative microscopy. ISME J; doi: 10.1038/s41396-021-00900-6
Jahn M.T., Markert S.M., Ryu T, Ravasi T, Stigloher C, Hentschel U, Moitinho-Silva (2016) Shedding light on cell compartmentation in the candidate phylum Poribacteria by high resolution visualisation and transcriptional profiling. Sci. Rep. 6; doi: 10.1038/srep35860
Bakkeren, E., Piskovsky, V., Lee, M.N.Y., Jahn, M.T., Foster KR 2025, Strain displacement in microbiomes via ecological competition. Nature Microbiology; doi: 10.1038/s41564-025-02162-w

