Complexes in Phage-infected Cells

Our research
Microbial viruses called phages infect bacteria, manipulate their cellular physiology, and finally lyse the cells. Many phages become an integral part of bacteria by integrating into their genome or coexisting in the population, while others can lyse an entire bacterial population. This leads to an ongoing conflict between phages and bacteria for billions of years. The cyclical lysis and resistance events between phages and bacteria have shaped evolution and invented unpredictable machines that can be exploited for breakthrough technologies such as CRISPR-Cas. The phage-host interface is the most diverse sequence space in the protein universe and here discoveries are to be made.
Our group is particularly interested in jumbo phages, which have genome sizes >200 kB and encode hundreds of factors of unknown function. These phages replicate rapidly, form cellular structures like in eukaryotic cells and have a huge repertoire of functions to take control of their host. This species-specific process requires adaptation and ingenuity on the side of the phage to overcome cellular immunity or stress. We aim to elucidate the mechanisms of phage host takeover and to functionally characterise their molecular mechanisms. We use multi-omics approaches to identify phage factors that interact with cellular production factories, and cryo-EM and biochemical assays to elucidate the structural and molecular mechanisms behind their roles. We aim to translate these factors and their mechanisms into novel therapies.
Complexes in phage-infected cells
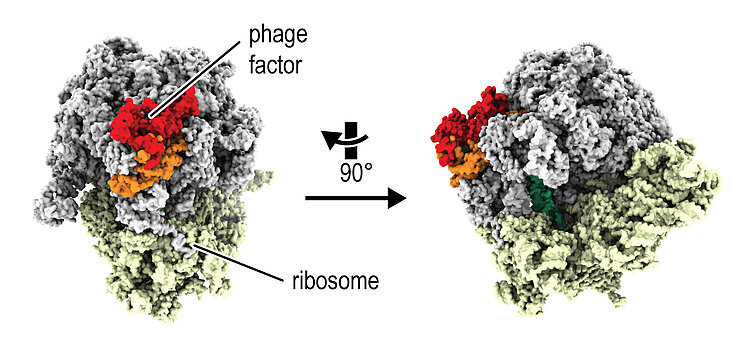
We characterise complexes between RNA, DNA and proteins in phage-infected cells. We have recently identified phage factors that directly target the protein production machinery, a megacomplex of proteins and RNA, the ribosome. To investigate the molecular mechanisms of these factors, we are using structural studies such as single-particle cryo-electron microscopy (cryo-EM) and biochemical assays. Our goal is to understand how phages modulate the stress response in protein production.
Cellular structures in phage-infected cells
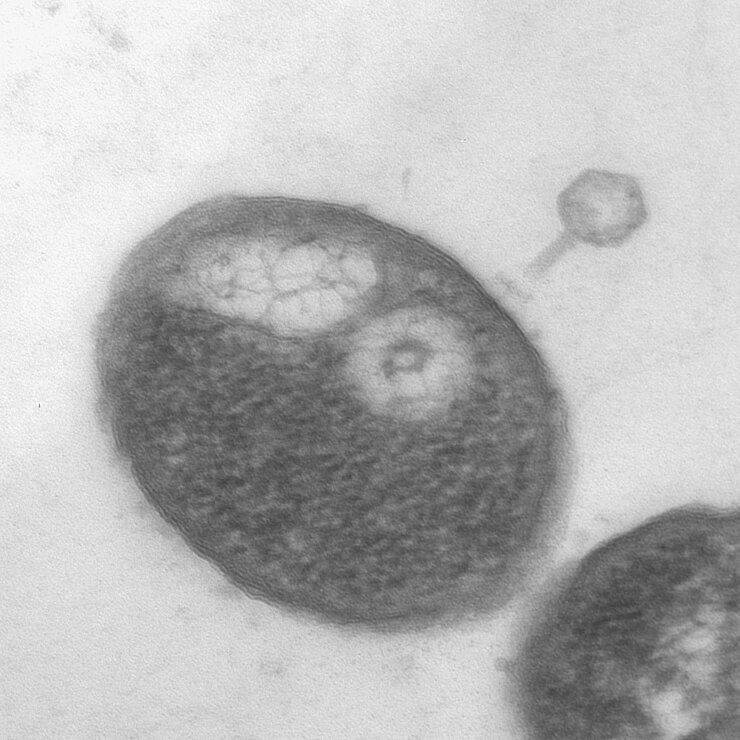
On first contact, phages inject their genetic payload into the host. Strikingly, some jumbo phages of the family Chimalliviridae form self-made cellular compartments that protect their payload but also serve as production facilities. For example, a proteinaceous phage nucleus is formed that is used to orchestrate phage production and assembly. We are curious to investigate how proteins and RNAs associate with phage-made cellular structures, adding a spatial layer of complexity to the phage replication cycle. In our studies, we look forward to discovering novel hidden cellular structures that could be used for artificial compartments in biotechnological and medical applications.
Our research
Microbial viruses called phages infect bacteria, manipulate their cellular physiology, and finally lyse the cells. Many phages become an integral part of bacteria by integrating into their genome or coexisting in the population, while others can lyse an entire bacterial population. This leads to an ongoing conflict between phages and bacteria for billions of years. The cyclical lysis and resistance events between phages and bacteria have shaped evolution and invented unpredictable machines that can be exploited for breakthrough technologies such as CRISPR-Cas. The phage-host interface is the most diverse sequence space in the protein universe and here discoveries are to be made.
Our group is particularly interested in jumbo phages, which have genome sizes >200 kB and encode hundreds of factors of unknown function. These phages replicate rapidly, form cellular structures like in eukaryotic cells and have a huge repertoire of functions to take control of their host. This species-specific process requires adaptation and ingenuity on the side of the phage to overcome cellular immunity or stress. We aim to elucidate the mechanisms of phage host takeover and to functionally characterise their molecular mechanisms. We use multi-omics approaches to identify phage factors that interact with cellular production factories, and cryo-EM and biochemical assays to elucidate the structural and molecular mechanisms behind their roles. We aim to translate these factors and their mechanisms into novel therapies.
Complexes in phage-infected cells
We characterise complexes between RNA, DNA and proteins in phage-infected cells. We have recently identified phage factors that directly target the protein production machinery, a megacomplex of proteins and RNA, the ribosome. To investigate the molecular mechanisms of these factors, we are using structural studies such as single-particle cryo-electron microscopy (cryo-EM) and biochemical assays. Our goal is to understand how phages modulate the stress response in protein production.
Cellular structures in phage-infected cells
On first contact, phages inject their genetic payload into the host. Strikingly, some jumbo phages of the family Chimalliviridae form self-made cellular compartments that protect their payload but also serve as production facilities. For example, a proteinaceous phage nucleus is formed that is used to orchestrate phage production and assembly. We are curious to investigate how proteins and RNAs associate with phage-made cellular structures, adding a spatial layer of complexity to the phage replication cycle. In our studies, we look forward to discovering novel hidden cellular structures that could be used for artificial compartments in biotechnological and medical applications.
Phages encode for the most diverse sequence space in the protein universe, full of unforeseen discoveries.
Dr Milan Gerovac
Dr. Milan Gerovac studied biochemistry at the Goethe University in Frankfurt, Germany. In his PhD thesis with Prof. Robert Tampé at Goethe University, he investigated translation control mechanisms and characterised complexes in protein production. He then joined the laboratory of Jörg Vogel at the Julius-Maximilians-University and the Helmholtz Institute for RNA-based Infection Research in Würzburg as a postdoctoral researcher, where he developed methods for the discovery and characterisation of RNA-binding proteins and gained experience in microbiology with various bacterial pathogens. He switched to studying phage biology by looking into complexes in phage-infected cells and developed a fascination with the dark protein space in phage genomes, discovering phage factors that target the protein production machine. In 2024, Gerovac started the junior research group Complexes in Phage-infected Cells (CPIC) at the HZI with the aim of discovering phage-based mechanisms that can be translated into novel therapeutic concepts to treat infections.
Team
We are always looking for new members to join our team and welcome you to contact us.
The laboratory is currently being set up and the team put together. Team members will be listed soon.
Selected publications
Gerovac M, Ðurica-Mitić S, Buhlmann L, Rech V, Zhu Y, Carien S, Popella L, Vogel J (2024)
Non-genetic messenger RNA silencing reveals essential genes in phage-host interplay.
bioRxiv, doi: 10.1101/2024.07.31.605949
Putzeys L & Wicke L & Brandão A, Boon M, Pires DP, Azeredo J, Vogel J, Lavigne R, Gerovac M (2024)
Exploring the transcriptional landscape of phage-host interactions using novel high-throughput approaches. Current Opinion in Microbiology 77, 102419. doi: 10.1016/j.mib.2023.102419
Gerovac M, Chihara K, Wicke L, Böttcher B, Lavigne R, Vogel J (2024)
Phage proteins target and co-opt host ribosomes immediately upon infection. Nature Microbiology 9, 787-800. doi: 10.1038/s41564-024-01616-x
Gerovac M, El Mouali Y, Kuper J, Kisker C, Barquist L, Vogel J (2020)
Global discovery of bacterial RNA-binding proteins by RNase-sensitive gradient profiles reports a new FinO domain protein. RNA 26, 1448-63. doi: 10.1261/rna.076992.120
*Heuer A & *Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kötter P, Berninghausen O, Becker T, Beckmann R & Tampé R (2017) Structure of the 40S–ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nature Structural & Molecular Biology 24, 453-60 doi: 10.1038/nsmb.3396
Technology Offers
The following technologies have been developed and patented by the research group "Complexes in Phage-infected Cells":
Novel ASO-Based mRNA Silencing Technology for Phage-Host Interaction Research
Newsroom
Are you interested in a bachelor, master or doctoral thesis? We are looking forward to your request!