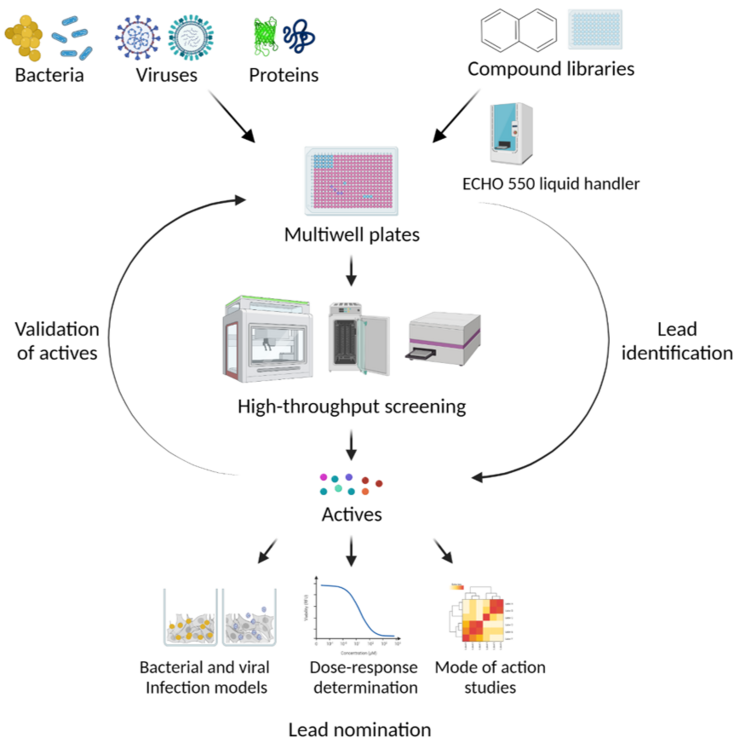
Compound Profiling and Screening

Our research
Infectious diseases occur when a potential pathogen manages to multiply uncontrollably in a host and damage organs or essential bodily functions. Treatment is successful when it is possible to stop the spread of the pathogen, minimize tissue and organ damage and, ideally, eliminate the pathogen. This can be achieved with the help of chemical substances that inhibit the biological processes essential for the damaging effect, survival and multiplication of the pathogen. Appropriate tests are required to identify the corresponding activities of substances. These tests must be representative of the respective infectious disease, i.e. they can, for example, address the vitality and reproductive capacity of the pathogen, but also the function and activity of individual components (proteins, nucleic acids, membranes) that are essential for successful infection.
We specialize in tests for infectious diseases caused by bacteria or viruses of biological safety levels 2 and 3. With the help of these tests, we can not only search specifically for substances against a certain infectious disease, but can also create biological activity profiles of substances provided to us by cooperation partners and thus contribute to the creation of broad structure-activity profiles for the more targeted development of anti-infectives.
In many cases, the test substances are only available in small quantities at the start of a screening project and many substances need to be tested for activity when addressing a new target structure or a new pathogen. For this purpose, we have established a technical infrastructure for which the availability of a few microliters of the substance solutions is sufficient and which allows the automated execution of tests. The test protocols are therefore being developed in such a way that they can be miniaturized and automated.
Current projects are aimed at
- Further development of inhibitors of the virulence factor α-haemolysin
- Discovery of substances with antiviral activity (Chikungunya virus, SARS-CoV-2, models for Marburg virus)
- Profiling the antibiotic activity of substances
Our research
Infectious diseases occur when a potential pathogen manages to multiply uncontrollably in a host and damage organs or essential bodily functions. Treatment is successful when it is possible to stop the spread of the pathogen, minimize tissue and organ damage and, ideally, eliminate the pathogen. This can be achieved with the help of chemical substances that inhibit the biological processes essential for the damaging effect, survival and multiplication of the pathogen. Appropriate tests are required to identify the corresponding activities of substances. These tests must be representative of the respective infectious disease, i.e. they can, for example, address the vitality and reproductive capacity of the pathogen, but also the function and activity of individual components (proteins, nucleic acids, membranes) that are essential for successful infection.
We specialize in tests for infectious diseases caused by bacteria or viruses of biological safety levels 2 and 3. With the help of these tests, we can not only search specifically for substances against a certain infectious disease, but can also create biological activity profiles of substances provided to us by cooperation partners and thus contribute to the creation of broad structure-activity profiles for the more targeted development of anti-infectives.
In many cases, the test substances are only available in small quantities at the start of a screening project and many substances need to be tested for activity when addressing a new target structure or a new pathogen. For this purpose, we have established a technical infrastructure for which the availability of a few microliters of the substance solutions is sufficient and which allows the automated execution of tests. The test protocols are therefore being developed in such a way that they can be miniaturized and automated.
Current projects are aimed at
- Further development of inhibitors of the virulence factor α-haemolysin
- Discovery of substances with antiviral activity (Chikungunya virus, SARS-CoV-2, models for Marburg virus)
- Profiling the antibiotic activity of substances
Prof Dr Ursula Bilitewskii
In order to develop therapies for fighting infections caused by new pathogens or resistant microorganisms not only we need new drugs but, more importantly, we need to identify new targets for these drugs to act on.

Ursula Bilitewski studied chemistry at the University of Münster, earning her Ph.D. at the Institute of Physical Chemistry. After spending another year at Münster as a post-doc, she moved to Braunschweig in 1988, where she became part of the HZI – which at that time was called the GBF.
In 1994, she habilitated in biochemistry at the Technische Universität Braunschweig and, since 2002, is an adjunct professor for biochemistry. Her initial research focus, which was on establishing microscale bioanalytical systems, has since shifted to drug research. Her work concentrates on molecular mechanisms and on identifying potential drug targets.
Since 2016, Bilitewski is head of the „Compound Profiling und Screening (COPS)“ research group.
Team








Selected Publications
Lettl, C.; Schindele, F.; Reza Mehdipour, A.; Witschel, M.; Steiner, T.; Ring, D.; Brack-Werner, R.; Stecher, B.; Eisenreich, W.; Bilitewski, U.; Hummer, G.; Fischer, W.; Haas, R. (2023) Selective killing of the human gastric pathogen Helicobacter pylori by mitochondrial respiratory complex I inhibitors, Cell Chem. Biol. DOI: 10.1016/j.chembiol.2023.04.003
Suerbaum S., Coombs N., Patel L., Pscheniza D., Rox K., Falk C., Gruber A.D., Kershaw O., Chhatwal P., Brönstrup M., Bilitewski U., Josenhans C.; Identification of Antimotilins, Novel Inhibitors of Helicobacter pylori Flagellar Motility That Inhibit Stomach Colonization in a Mouse Model; mBio (2022) DOI: 10.1128/mbio.03755-21
Sabrina Mühlen, Viktor A. Zapolskii, Ursula Bilitewski, Petra Dersch, Identification of translocation inhibitors targeting the type III secretion system of enteropathogenic Escherichia coli, Antimicrob. Agents Chemotherap. (2021) DOI: 10.1128/AAC.00958-21
Brennecke P, Rasina D, Aubi O, Herzog K, Landskron J, Cautain B, Vicente F, Quintana J, Mestres J, Stechmann B, Ellinger B, Brea J, Kolanowski JL, Pilarski R, Orzaez M, Pineda-Lucena A, Laraia L, Nami F, .. Bilitewski U, Brönstrup M, …Genilloud O, .. Philip Gribbon, EU-OPENSCREEN: A Novel Collaborative Approach to Facilitate Chemical Biology, SLAS Discovery 24 (2019) 398 – 413 DOI: 10.1177/2472555218816276
A. Heintz-Buschart, H. Eickhoff, E. Hohn, U. Bilitewski, Identification of inhibitors of yeast-to-hyphae transition in Candida albicans by a reporter screening assay, J. Biotechnol. 164 (2013) 137 - 142 DOI: 10.1016/j.jbiotec.2012.12.004
Publications
Projects
Inhibition of the activity of α-hemolysin of Staphylococcus aureus
α-hemolysin is one of the proteins of S. aureus that is secreted by the bacteria and leads to damage of human cells by forming pores in the membrane of the cells. In humans, it particularly attacks cells of the immune system and epithelial and endothelial cells and is the decisive virulence factor in pneumonia and skin infections caused by S. aureus. For a screening campaign at the Lead Discovery Center (LDC) in Dortmund with 180,000 substances, we developed the screening test with the recombinant protein and provided test substances. To validate the activity of the inhibitors found, we established infection models in which the lung epithelial cell A549 was specifically infected with S. aureus. Using this model, we were also able to characterize the pathogenic properties of more than 100 isolates from patients. The added inhibitors were able to prevent damage to the cells by α-hemolysin and also the formation of inflammatory markers (cytokines). Special cell culture models in which infection occurs at the boundary layer between air and cell culture (air-liquid interface cultures) are being established for even more extensive investigations under conditions close to infection. We are also establishing diagnostic methods to quantify α-hemolysin in patient samples, as the presence of significant α-hemolysin concentrations cannot be inferred from the presence of S. aureus alone when inhibitors are used prophylactically.
Duration: since 2015
Funded by: Helmholtz Validation Fund, Pre4D program, CARB-X, DZIF
Antiviral agents
Chikungunya - Virus
The chikungunya virus is transmitted by mosquitoes, causes severe muscle and joint pain and headaches and is accompanied by a high fever. Newborns, the elderly and people with chronic illnesses are at risk of severe cases. Originally, the virus was mainly found in South East Asia and Africa, but has been spreading worldwide for some time, including to Europe, in line with the spread of mosquitoes. There is currently no specific treatment.
In collaboration with Prof. J. Diez, Barcelona and funded by the European Commission, we screened the EU-OPENSCREEN substance library of approx. 100,000 substances for substances that inhibit the replication of the Chikungunya virus. We were able to use a reporter virus that contained luciferase in the genome so that the luciferase activity was a measure of the number of virus particles. We were also able to quantify the cytopathic effects caused by the virus.
To achieve a sufficiently high substance throughput and to minimize substance consumption, 384well microtiter plates were used for the assay. The substance plates were prepared by transferring 100 nL of the substance solutions through the Echo®550 acoustic dispenser. The actual infection assay was performed in the S3** laboratory. The same protocol was also used to investigate the anti-Chikungunya activity of natural products or other chemically synthesized substances. Some very promising substances were identified, which were further investigated in a medicinal chemistry project (funded by EU-OPENSCREEN).
Metabolome studies were carried out to investigate the interaction between the virus and host cells in more detail, and CRISPR-Cas9 technology was established. The aim is to identify new targets for active substances that can be addressed by specific tests.
Duration of the project: since 2020
Funded by: European Commission through a demonstration project of the EU-OPENSCREEN research infrastructure and through a cooperation project within the framework of cooperation with Southeast Asia;
Fellowship from the Alexander von Humboldt Foundation
SARS-CoV-2
The outbreak of the Covid-19 pandemic triggered global efforts to find active substances to treat this disease, as most of the known antiviral drugs did not have a satisfactory effect against SARS-CoV-2. We used the EU-OPENSCREEN substance library (approx. 100,000 substances) in screening campaigns.
In cooperation with Prof. E. Kierzek, Polish Academy of Sciences, Poznan and funded by the European consortium of research infrastructures ISIDORe, a test was established that can be used to search for intercalators in conserved RNA secondary structures in a largely automated manner. After determining the effective concentration ranges (IC50 values), the activity of the substances is checked in the infection assay with SARS-CoV-2. An automated test facility is also available in the S3 laboratory for this purpose.
Duration: 2020 - 2025
Funded by: European Commission through the European Research Infrastructures Network ISIDORe
Preparing for future pandemics: COMBINE
The Marburg virus will be used to develop strategies for the prevention and treatment of future viral infections. The project is coordinated by Prof. Christian Sieben (HZI). The task of the COPS working group / CBIO department is to search for inhibitors of interactions between phosphatidylserine, viral glycoproteins and viral proteins with their respective receptors on the surface of host cells. These inhibitors are intended to prevent the uptake of virus particles into cells and thus the infection of the cells. The EU-OPENSCREEN library as well as natural products and target-specific libraries are used as substance libraries. In order to achieve a sufficient throughput of substances, the aim is to automate the tests as far as possible. Further information can be found at www.combine-MARV.eu.
Duration: 2025 - 2029
Funded by: European Commission
Technology offers
Screening campaigns
The Department of Chemical Biology (CBIO) has equipment available for the automated performance of biochemical / microbiological / cell biological tests. These are integrated systems with a pipetting robot (i7 or i5), automatic incubator (Cytomat), multi-mode microplate reader (Synergy, Biotek) and dispenser (Multidrop). There are two very similar systems, one of which is located in a biosafety level 3 environment and the other in biosafety level 2 laboratories, so that work can also be carried out directly with the relevant pathogens. The CBIO department is therefore one of the official screening centers of the European research infrastructure EU - OPENSCREEN.
We offer the adaptation of test protocols to the systems and the implementation of screening campaigns, for example with substance libraries of the CBIO department (approx. 40,000 substances) and / or the European research infrastructure EU - OPENSCREEN (approx. 100,000 substances) and establish the necessary contacts. The facility is suitable for investigating the effect not only of chemical substances, but also of proteins, nucleic acids or other agents with a potentially inhibitory or activating effect. The only requirement is that the tests can be carried out in microtiter plates.
We produce the microtiter test plates required for the screening campaigns with the Echo®550 acoustic dispenser, if this is possible by transferring solutions of chemical substances in DMSO. By using the Echo®550, volumes smaller than 1 µL can be easily transferred from a mother plate to the test plate. Concentration dependencies can also be investigated very easily by transferring different volumes. This technology minimizes the need for test substance. In addition, no pipette tips are required as the transfer is carried out using acoustic energy.
If necessary, we also support external screening campaigns by producing substance plates.
All screening campaigns also include the administration and evaluation of the data so that the screening data can be assigned to chemical substances.
Profiling the antibiotic activity of chemical substances
We offer to determine the basic biological activities of chemical substances as part of research collaborations. These are initially studies on antibiotic effects, i.e. on an inhibitory influence on the growth of bacteria. For initial orientation, we use Staphylococcus aureus as the relevant representative of Gram-positive bacteria and Escherichia coli for Gram-negative bacteria. In order to exclude the possibility that the lack of activity against Escherichia coli is caused by efficient export of substances from the bacteria, we also use the ΔTolC mutant of E. coli, as one of the most important substance export systems is inactivated in it. In the event of promising activities, further bacteria can then be included in the investigations.
In addition to the tests for antibiotic effects, we also characterize the substances for their effect on the vitality of animal cells, as this also provides information on possible applications of the observed effects.
As we can use the Echo®550 acoustic dispenser to handle substances that are soluble in DMSO, we only need around 10 µL of the substance solutions for the initial tests.
As a screening center of the European research infrastructure EU - OPENSCREEN, we have examined the complete substance library of more than 100,000 substances available there with regard to antibiotic activity against the four Gram-negative bacteria from the ESKAPE panel, namely Escherichia coli, Acinetobacter baumanii, Klebsiella pneumoniae and Pseudomonas aeruginosa.