Core Facility for Translational Proteomics

Our Expertise
The Core Facility for Translational Proteomics (CFTP) provides proteome-scaled analyses as full service as well as in collaborative projects. We combine expertise in protein biochemistry, mass spectrometry (MS) and biostatistics matching the demands in academical and translational research.
The Core Facility is equipped with modern mass spectrometry with ion mobility for untargeted and targeted proteome studies (DDA, DIA, PRM). Data analysis is conducted using a combination of commercial, academic, and in-house software solutions and databases, ensuring project-specific data mining and management. The established workflows allow the identification of proteins and post-translational mechanisms that are key regulators in infections. By this, CFTP contributes to the discovery of novel drug targets in host-pathogen interaction networks that coordinate the course of infections.
Our Expertise
The Core Facility for Translational Proteomics (CFTP) provides proteome-scaled analyses as full service as well as in collaborative projects. We combine expertise in protein biochemistry, mass spectrometry (MS) and biostatistics matching the demands in academical and translational research.
The Core Facility is equipped with modern mass spectrometry with ion mobility for untargeted and targeted proteome studies (DDA, DIA, PRM). Data analysis is conducted using a combination of commercial, academic, and in-house software solutions and databases, ensuring project-specific data mining and management. The established workflows allow the identification of proteins and post-translational mechanisms that are key regulators in infections. By this, CFTP contributes to the discovery of novel drug targets in host-pathogen interaction networks that coordinate the course of infections.
Team




We provide service for:
- Quality and purity control of biologicals (e.g. antibodies)
- Protein identification (gel free, gel based)
- Quantitative proteome analytics
- In vivo: SILAC & BONCAT
- Ex vivo: iTRAQ & TMT
- Molecular phenotyping of tissues, organoids and cells
- Phenotyping of mutant cells (CRISPR-Cas9)
- Biomarker studies (clinical proteomics, cohort studies)
- Interactome studies (IP-MS)
- Post-translational modification (PTM) analyses (e.g. phosphorylation, glycosylation, ubiquitination)
- Drug target identifications
- De novo peptide sequencing and protein analytics (ETD/MSn)
Equipment:

CFTP operates the following modern MS workflows:
Orbitrap (ETD-Fusion & Velos Pro*) mass spectrometer combined with HPLC system (Ultimate 3000, Waters NanoAcquity*) for biologicals, labile PTMs and quantitative proteomics.
Ion-mobility mass spectrometry (timsTOF Pro) (DDA; DIA; PRM) combined with high throughput LC system (Evosep One) provides a dedicated workflow for clinical proteomics and epidemiological cohort studies such as in plasma proteomics.
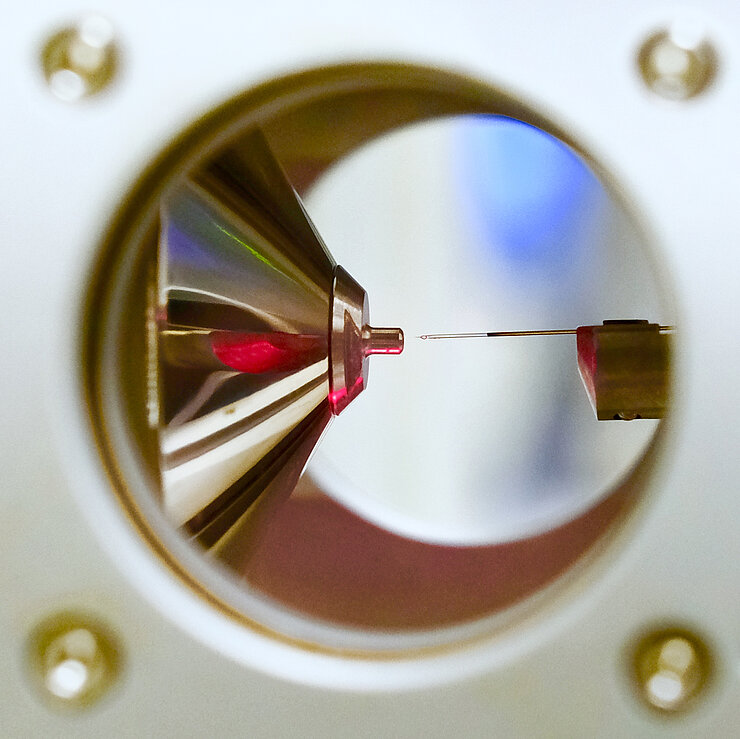
Orbitrap Exploris 480* mass spectrometer with the FAIMS Pro Duo, ultra-sensitive workflow for low abundant proteins and protein networks (including single cell proteomics, spatial proteomics).
Multi-Core Workstation (96-Core CPU) for advanced MS data software packages and in-house data base searches (including Proteome Discoverer, MaxQuant, Peaks Studio, Spectronaut, Spectrodive).
* with HZI proteome research groups as collaborative service
Research & Development:
We explicitly enable research and development (R&D) in direct collaboration with scientific groups whose research questions require specific protein and proteome analysis. Our mission is to enable scientific discoveries in infection research by developing unique analytical pipelines. We have a particular interest in interdisciplinary studies that establish joint activities with other institutions and central facilities.
Our main interests are:
- Glycoproteome strategies
- Spatial Proteomics (including laser dissection aided sampling)
- Single cell Proteomics (including CELLENION Technology)
- Protein enrichment by functionalized nanoparticles
- Multi-Omics strategies
5 selected Publications:
Shekhar A, Di Lucrezia R, Jerye K, Korotkov VS, Harmrolfs K, Rox K, Weich HA, Ghai I, Delhommel F, Becher I, Degenhart C, Fansa E, Unger A, Habenberger P, Klebl B, Lukat P, Schmelz S, Henke S, Borgert S, Lang JC, Sasse F, Diestel R, Richter C, Schneider-Daum N, Hinkelmann B, Niemz J, Lehr CM, Jänsch L, Huehn J, Alm R, Savitski M, Welte T, Hesterkamp T, Sattler M, Winterhalter M, Blankenfeldt W, Medina E, Bilitewski U, Dinkel K, Brönstrup M (2025) Highly potent quinoxalinediones inhibit α-hemolysin and ameliorate Staphylococcus aureus lung infections. Cell Host Microbe 25: S1931-3128(25)00089-7
Ayala-García P, Herrero-Gómez I, Jiménez-Guerrero I, Otto V, Moreno-de Castro N, Müsken M, Jänsch L, van Ham M, Vinardell JM, López-Baena FJ, Ollero FJ, Pérez-Montaño F, Borrero-de Acuña JM (2025) Extracellular Vesicle-Driven Crosstalk between Legume Plants and Rhizobia: The Peribacteroid Space of Symbiosomes as a Protein Trafficking Interface. J Proteome Res 24(1):94-110
Garza AP, Wider-Eberspächer E, Morton L, van Ham M, Pállinger É, Buzás EI, Jänsch L, Dunay IR (2024) Proteomic analysis of plasma-derived extracellular vesicles: pre- and postprandial comparisons. Sci Rep. 14(1):23032
Jerye K, Lüken H, Steffen A, Schlawis C, Jänsch L, Schulz S, Brönstrup M (2024) Activity-Based Protein Profiling Identifies Protein Disulfide-Isomerases as Target Proteins of the Volatile Salinilactones. Adv Sci (Weinh) 11(18):e2309515
Schöl M, Schempp R, Hennig T, Wigger D, Schumacher F, Kleuser B, Stigloher C, van Ham M, Jänsch L, Schneider-Schaulies S, Dölken L, Avota E (2024) Dynamic changes in the proximitome of neutral sphingomyelinase-2 (nSMase2) in TNFα stimulated Jurkat cells. Front Immunol 15:1435701
Contact and Materials:
Would you like to request a Service? Please contact us by email with your request and use this form:
For consultation or specific questions, please contact:
- Prof. Dr. Lothar Jänsch (Consulting, Clinical and Cellular Proteomics)
- Dr. Dominik Körner (Phenotyping, Interactomics, Service, Quotations)
- Dr. Tina Rietschel (PTM-Proteomics, Glycoproteomics) (CPRO)
- Dr. Marco van Ham (Single Cell and Spatial Proteomics) (CPRO)
- Prof. Dr. Frank Klawonn (Bioinformatics, Statistics) (CPRO)
- Prof. Dr. Susanne Engelmann (Microbiology, Metaproteomes, Dark Proteome) (MPRO)