Dynamics of Respiratory Infections

Our research
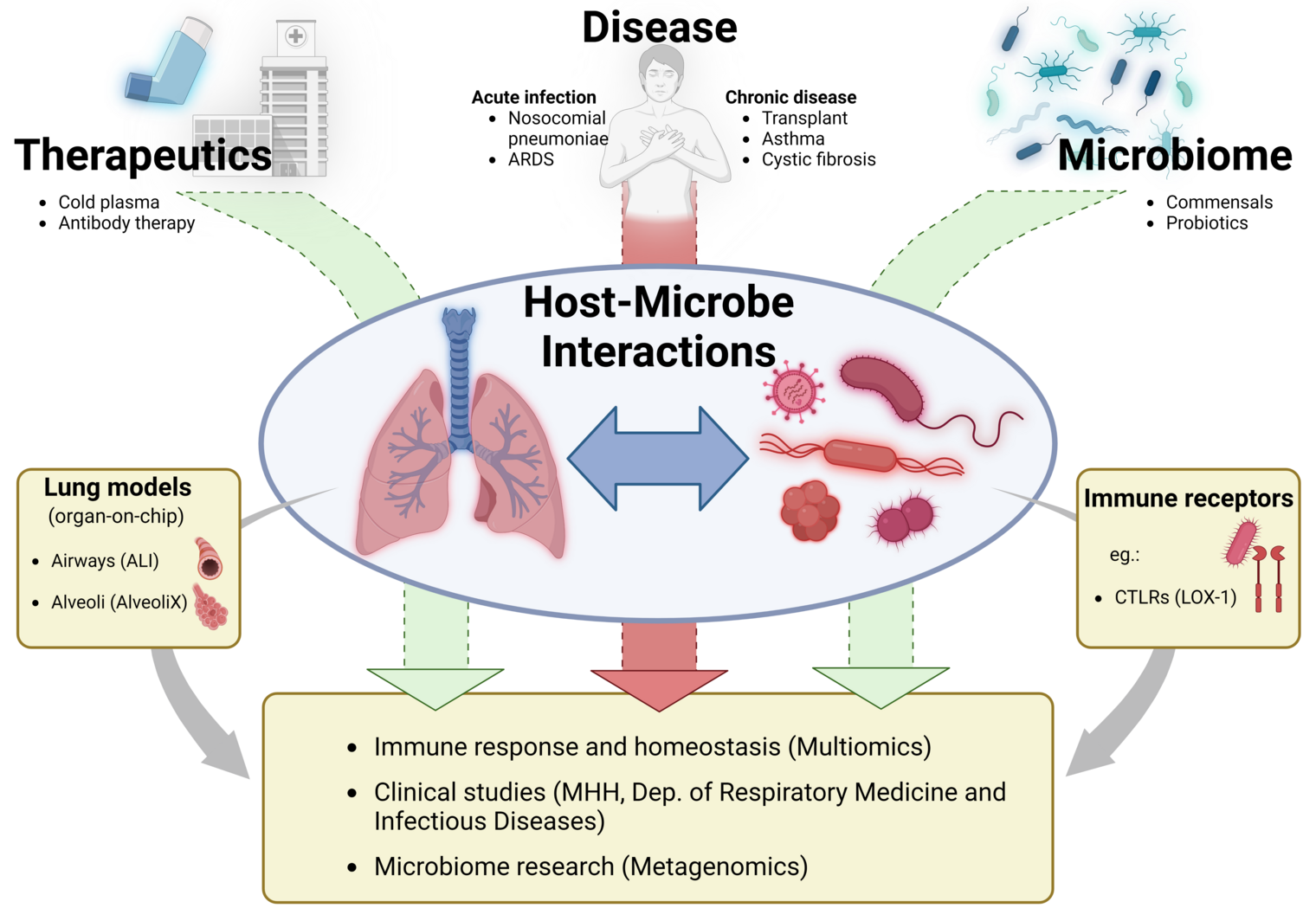
The research group “Dynamics of Respiratory Infections” (DINF) focuses on studying pathogen-host interactions in the respiratory tract, with a particular emphasis on bacterial pathogens such as Staphylococcus aureus and Streptococcus pneumoniae, as well as fungal pathogens of the lung, including Aspergillus species. Our research investigates receptor-mediated mechanisms by which these pathogens interact with the pulmonary epithelium and alveolar macrophages. Additionally, we analyze the role of non-pathogenic lung colonizers, which influence pathogen behavior and modulate the host’s innate immunity by altering immune signaling pathways.
Our research
The research group “Dynamics of Respiratory Infections” (DINF) focuses on studying pathogen-host interactions in the respiratory tract, with a particular emphasis on bacterial pathogens such as Staphylococcus aureus and Streptococcus pneumoniae, as well as fungal pathogens of the lung, including Aspergillus species. Our research investigates receptor-mediated mechanisms by which these pathogens interact with the pulmonary epithelium and alveolar macrophages. Additionally, we analyze the role of non-pathogenic lung colonizers, which influence pathogen behavior and modulate the host’s innate immunity by altering immune signaling pathways.
Prof Dr med Hortense Slevogt
A better understanding of the host-microbe interactions in the lung may help us to develop novel therapeutic strategies for a variety of respiratory infections.

Prof Dr med Hortense Slevogt has established herself as a distinguished researcher and physician in the fields of respiratory infections and immunology. Her academic journey began with a degree in medicine at Freie Universität Berlin, during which she developed a particular interest in infectious diseases and their impact on respiratory health. Driven by her passion for understanding the intricate workings of the human immune system, she pursued further specialization in Internal Medicine and Pulmonology, becoming a recognized expert in the field.
In 2009, Prof Slevogt's achievements were acknowledged through the completion of her habilitation in Internal Medicine at Charité - Universitätsmedizin Berlin. This significant milestone marked her expertise and profound understanding of the intricate nature of severe infections. A few years later, in 2011, Prof. Slevogt assumed the leadership of the Host Septomics Group at Universitätsklinikum Jena.
In 2022, she obtained the W3-Professorship at Medizinische Hochschule Hannover (MHH) and became the Head of the Research Group "Respiratory Infection Dynamics" (DINF) at the Helmholtz Centre for Infection Research (HZI) in Braunschweig. Her role entails spearheading translational projects that involve collaborations with esteemed academic and medical partners at the MHH. Through these endeavors, Dr Slevogt investigates host-pathogen interactomes and elucidates the mechanisms underlying immune responses and their control.
In parallel to her research pursuits, Dr Slevogt remains dedicated to clinical practice, ensuring that her findings directly benefit patients. She serves as the Senior Consultant in Clinical Infectiology at the Department of Respiratory Medicine and Infectious Diseases (MHH), contributing her expertise to the diagnosis and treatment of respiratory infections.










Selected Publications
Klassert TE #, Goyal S, Stock M, Driesch D, Hussain A, Berrocal-Almanza LC, Myakala R, Sumanlatha G, Valluri V, Ahmed N, Schumann RR, Flores C, Slevogt H §. (2018). AmpliSeq Screening of Genes Encoding the C-Type Lectin Receptors and Their Signaling Components Reveals a Common Variant in MASP1 Associated with Pulmonary Tuberculosis in an Indian Population. Front Immunol. 9:242. 10.3389/fimmu.2018.00242.
Lehmann R, Müller MM, Klassert TE, Driesch D, Stock M, Heinrich A, Conrad T, Moore C, Schier UK, Guthke R, Slevogt H §. (2018). Differential regulation of the transcriptomic and secretomic landscape of sensor and effector functions of human airway epithelial cells. Mucosal Immunol. 11(3):627-642. 10.1038/mi.2017.100.
Klassert TE #, Leistner R, Zubiria-Barrera C, Stock M, López M, Neubert R, Driesch D, Gastmeier P, Slevogt H §. (2021). Bacterial colonization dynamics and antibiotic resistance gene dissemination in the hospital environment after first patient occupancy: a longitudinal metagenetic study. Microbiome. 9(1):169. 10.1186/s40168-021-01109-7.
Klassert TE #, Zubiria-Barrera C, Neubert R, Stock M, Schneegans A, López M, Driesch D, Zakonsky G, Gastmeier P, Slevogt H §, Leistner R. (2022). Comparative analysis of surface sanitization protocols on the bacterial community structures in the hospital environment. Clin Microbiol Infect. 28(8):1105-1112. 10.1016/j.cmi.2022.02.032.
Müller MM, Baldauf C, Hornischer S, Klassert TE, Schneegans A, Behnert A, Pletz MW, Hagel S, Slevogt H §. (2023). Staphylococcus aureus induces tolerance in human monocytes accompanied with expression changes of cell surface markers. Front Immunol. 14:1046374. 10.3389/fimmu.2023.1046374.
Projects
- The influence of microbial commensal-pathogen interactions on shaping the immunological environment of the human respiratory tract (RESIST)
- PlasmaCare: Analysis of the safety of cold plasma application on human lung epithelium using complex cell culture models.
- The interplay of Candida albicans and Aspergillus fumigatus induced signals in receptor-mediated modulation of antifungal immune responses
- Development of a novel test kit for the diagnosis of pneumocystis jirovecii infection