Genome Architecture and Evolution of RNA Viruses

Our Research
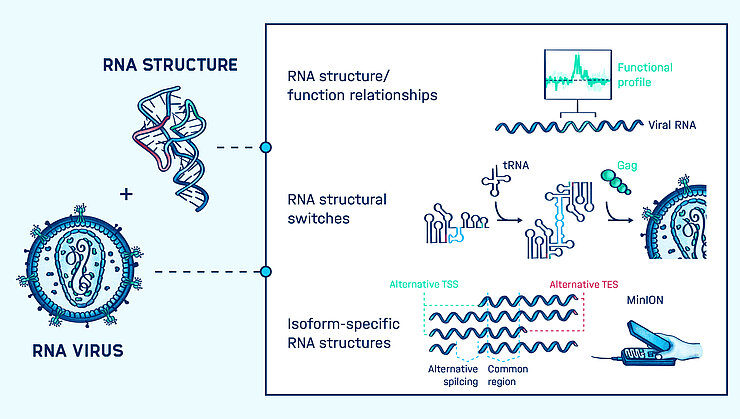
RNA is a functionally versatile molecule. It can specifically interact with proteins, small molecules, and base pair to other nucleic acids with exquisite precision. RNA viruses exploit this functional versatility at almost every stage of their replication cycle, using non-coding RNA elements to influence splicing, protein translation, evasion of host cell defenses, viral evolution and accessibility towards drug binding. Consequently, non-coding RNA represents an extremely attractive target for antiviral intervention, with the potential to revolutionize the treatment of infectious disease.
We use an integrative structural, functional and evolutionary approach to discover and mechanistically characterize non-coding RNA structures involved in viral replication and evolution. As in the protein world, it is often the higher order structure of the RNA, rather than primary sequence, that determines its function. Currently, how RNA structure drives diverse biological functions is not yet fully understood. Moreover, RNA readily undergoes structural changes, allowing it to switch between different functions, between different on/off states, or to adopt specific folds in different environments or in the presence of ligands. RNA dynamics have traditionally frustrated RNA structural characterization by biochemical and biophysical approaches. Our research focuses on unravelling the relationship between RNA structure and function, and we are actively working on new methods to investigate RNA structural dynamics. In the long term, we plan to use this knowledge to rationally develop small molecule drugs that interfere with RNA structure as a novel antiviral strategy.
We are also interested in how RNA structure constrains viral evolution. Retroviruses, such as HIV, package two copies of their RNA genome into each virion leading to recombination (template switching) and the formation of genome chimeras during replication. Another widespread strategy, seen in rotaviruses and influenza viruses, is genome segmentation leading to reassortment. Reassortment and recombination are non-random processes that are known to depend on RNA sequence and structure, but the underlying mechanisms are poorly understood. We study these mechanisms with the goal of improving disease prevention and control strategies. At the population level, we hope to understand the emergence of novel viral strains, such as how potentially pandemic influenza arises from genetic reassortment in pigs or birds. At an individual level, we want to understand how RNA structure leads to immune evasion and the generation of multiple drug resistant viruses. Through our fundamental research we seek to rationally manipulate recombination and reassortment for the development of safer gene therapy vectors, as well as powerful new vaccine platforms.
Our Research
RNA is a functionally versatile molecule. It can specifically interact with proteins, small molecules, and base pair to other nucleic acids with exquisite precision. RNA viruses exploit this functional versatility at almost every stage of their replication cycle, using non-coding RNA elements to influence splicing, protein translation, evasion of host cell defenses, viral evolution and accessibility towards drug binding. Consequently, non-coding RNA represents an extremely attractive target for antiviral intervention, with the potential to revolutionize the treatment of infectious disease.
We use an integrative structural, functional and evolutionary approach to discover and mechanistically characterize non-coding RNA structures involved in viral replication and evolution. As in the protein world, it is often the higher order structure of the RNA, rather than primary sequence, that determines its function. Currently, how RNA structure drives diverse biological functions is not yet fully understood. Moreover, RNA readily undergoes structural changes, allowing it to switch between different functions, between different on/off states, or to adopt specific folds in different environments or in the presence of ligands. RNA dynamics have traditionally frustrated RNA structural characterization by biochemical and biophysical approaches. Our research focuses on unravelling the relationship between RNA structure and function, and we are actively working on new methods to investigate RNA structural dynamics. In the long term, we plan to use this knowledge to rationally develop small molecule drugs that interfere with RNA structure as a novel antiviral strategy.
We are also interested in how RNA structure constrains viral evolution. Retroviruses, such as HIV, package two copies of their RNA genome into each virion leading to recombination (template switching) and the formation of genome chimeras during replication. Another widespread strategy, seen in rotaviruses and influenza viruses, is genome segmentation leading to reassortment. Reassortment and recombination are non-random processes that are known to depend on RNA sequence and structure, but the underlying mechanisms are poorly understood. We study these mechanisms with the goal of improving disease prevention and control strategies. At the population level, we hope to understand the emergence of novel viral strains, such as how potentially pandemic influenza arises from genetic reassortment in pigs or birds. At an individual level, we want to understand how RNA structure leads to immune evasion and the generation of multiple drug resistant viruses. Through our fundamental research we seek to rationally manipulate recombination and reassortment for the development of safer gene therapy vectors, as well as powerful new vaccine platforms.
Understanding the complex relationship between RNA structure and function will allow viral RNA genomes to be targeted in novel antiviral strategies

Redmond Smyth carried out his undergraduate degree in Natural Sciences at the University of Cambridge (UK), specializing in Virology and Immunology. He then moved to the Burnet Institute, Melbourne (Australia) for his PhD, where he investigated the mechanisms of HIV-1 genetic diversity in its natural target cells. His PhD research pointed to the importance of RNA structure in regulating viral infection processes, which is why he then moved to Strasbourg (France) for his post-doctoral research to train as an RNA biochemist. Here, he worked on understanding the mechanisms leading to the incorporation of the HIV-1 genomic RNA into viral particles. In 2015 he was recruited to the CNRS as a chargé de recherche (CR2), and in 2018, with support from the Helmholtz Association, he started his own research group at the Helmholtz Institute for RNA-based Infection Research (HIRI).
Selected Publications
Smyth RP§*, Smith MR*, Jousset AC, Despons L, Laumond G, Decoville T, Cattenoz P, Moog C, Jossinet F, Mougel M, Paillart JC, von Kleist M§, Marquet R§ (2018)
In cell mutational interference mapping experiment (in cell MIME) identifies the 5′ polyadenylation signal as a dual regulator of HIV-1 genomic RNA production and packaging
Nucleic Acids Research 46(9), e57
Mailler E, Bernacchi S, Vivet-Boudou V, Paillart JC, Marquet R, Smyth RP§ (2016)
The life-cycle of the HIV-1 gag-RNA complex
Viruses 8(9)
Smyth RP§, Despons L, Huili G, Bernacchi S, Hijnen M, Mak J, Jossinet F, Weixi Li, Paillart Jean-Christophe, von Kleist M§, Marquet R§ (2015)
Mutational Interference Mapping Experiment (MIME) for studying the relationship between RNA structure and function
Nature Methods 12(9):866-72
Abd El-Wahab EW*, Smyth RP*, Mailler E, Bernacchi S, Vivet-Boudou V., Hijnen M, Jossinet F, Mak J, Paillart JC, Marquet R (2014)
Specific recognition of the HIV-1 genomic RNA by the Gag precursor
Nature Communications 5:4304.
Gavazzi C, Yver M, Isel C, Smyth RP, Rosa-Calatrava M, Lina B, Moules V, Marquet R (2013)
A functional sequence-specific interaction between influenza A virus genomic RNA segments
PNAS 110(41):16604-9.