Host-Pathogen-Microbiota Interactions

Our Research
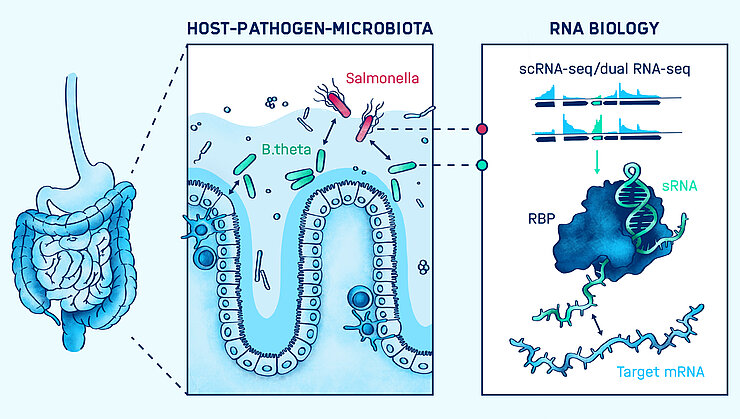
Bacterial infections of mammalian hosts are arguably among the most complex biological processes, often comprising a multitude of interacting organisms from different kingdoms. How do bacterial pathogens promote infection and what defense mechanisms do they have to overcome in order to colonize? What molecular mechanisms manifest the protective role of the microbiota against pathogenic attack? And what is the role of noncoding RNAs in host-microbiota-pathogen crosstalk? These and related questions are addressed in our group. Using cutting-edge RNA-sequencing-based techniques, our research centers on the identification and functional characterization of noncoding RNA molecules in pathogens, microbiota members, and the host, to identify those RNAs that may serve as biomarkers for diagnosis or as therapeutic targets in the future. In addition to contributing to the field by the development of novel RNA-seq-based technologies for complex infection settings, we aim to increase the knowledge about functions of regulatory RNA molecules and RNA-binding proteins in bacterial pathogenesis and symbiosis, by gaining biological insights from mechanistic studies.
Our Research
Bacterial infections of mammalian hosts are arguably among the most complex biological processes, often comprising a multitude of interacting organisms from different kingdoms. How do bacterial pathogens promote infection and what defense mechanisms do they have to overcome in order to colonize? What molecular mechanisms manifest the protective role of the microbiota against pathogenic attack? And what is the role of noncoding RNAs in host-microbiota-pathogen crosstalk? These and related questions are addressed in our group. Using cutting-edge RNA-sequencing-based techniques, our research centers on the identification and functional characterization of noncoding RNA molecules in pathogens, microbiota members, and the host, to identify those RNAs that may serve as biomarkers for diagnosis or as therapeutic targets in the future. In addition to contributing to the field by the development of novel RNA-seq-based technologies for complex infection settings, we aim to increase the knowledge about functions of regulatory RNA molecules and RNA-binding proteins in bacterial pathogenesis and symbiosis, by gaining biological insights from mechanistic studies.
Prof Dr Alexander Westermann
Infection is a complex interplay of a pathogen, its host, and the resident microbiota that we can only fully understand – and eventually treat – once we consider the role of each player in this process.

Alexander Westermann studied Molecular Biosciences at the University of Heidelberg (Germany) and worked as a visiting scholar in 2009 at UC Berkeley (California, USA). He obtained his PhD and worked as a PostDoc in the lab of Prof. Jörg Vogel at the Institute of Molecular Infection Biology (IMIB) in Würzburg. In 2017 and 2018, he was a visiting researcher in the labs of Prof. Andreas Bäumler (UC Davis, USA) and Prof. David Holden (Imperial College London, UK). Since March 2018, he is a Junior Professor at the IMIB and leads the HOPI group at the HIRI.
Selected Publications
Ryan D, Bornet E, Prezza G, Varshini Alampalli S, Franco de Carvalho T, Felchle H, Ebbecke T, Hayward R, Deutschbauer AM, Barquist L, Westermann AJ (2024)
An expanded transcriptome atlas for Bacteroides thetaiotaomicron reveals a small RNA that modulates tetracycline sensitivity.
Nature Microbiology 9(4):1130-1144.
Prezza G, Liao C, Reichardt S, Beisel CL, Westermann AJ (2024)
CRISPR-based screening of small RNA modulators of bile susceptibility in Bacteroides thetaiotaomicron.
PNAS 121(6):e2311323121.
Westermann AJ*,+, Vogel J*,+ (2021)
Cross-species RNA-seq for deciphering host-microbe interactions.
Nature Reviews Genetics 22(6):361-378.
Ryan D, Jenniches L, Reichardt S, Barquist L, Westermann AJ (2020)
A high-resolution transcriptome map identifies small RNA regulation of metabolism in the gut microbe Bacteroides thetaiotaomicron.
Nature Communications 11(1):3557.
Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J
Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions
Nature 2016, 529(7587): 496-501