Integrative Informatics for Infection Biology

Our Research
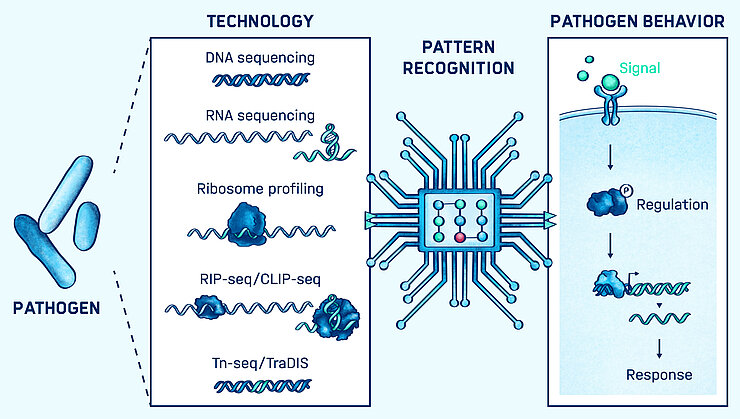
High-throughput functional genomics technologies are providing unprecedented views of cellular behavior at genome scale. Understanding how bacterial and host cells respond to complex processes like infection increasingly requires the integration and unified interpretation of these diverse technologies.
We are using modern data science technologies, including visualization, machine learning, and statistical modeling to extract biological insight from high-throughput genomic and post-genomic data. We have particular interests in using these technologies to understand the effects of RNA-based regulation in bacteria, and non-coding RNA’s role in host-pathogen interactions and the evolution of pathogens.
Our Research
High-throughput functional genomics technologies are providing unprecedented views of cellular behavior at genome scale. Understanding how bacterial and host cells respond to complex processes like infection increasingly requires the integration and unified interpretation of these diverse technologies.
We are using modern data science technologies, including visualization, machine learning, and statistical modeling to extract biological insight from high-throughput genomic and post-genomic data. We have particular interests in using these technologies to understand the effects of RNA-based regulation in bacteria, and non-coding RNA’s role in host-pathogen interactions and the evolution of pathogens.
Jun Prof Dr Lars Barquist
High-throughput technologies are changing the way biology is done, requiring the development of new computational and statistical tools. We aim to harness these technologies to bring new insights into bacterial pathogenesis.

Lars Barquist studied biomathematics at Rutgers University (New Jersey, USA) before working in the department of bioengineering at the University of California, Berkeley. He received a PhD from the University of Cambridge (UK) in 2014 for work on comparative functional pathogen genomics at the Wellcome Trust Sanger Institute. From 2014 to 2016 he was supported by an Alexander von Humboldt research fellowship at the Institute for Molecular Infection Biology at the University of Würzburg.
Selected Publications
Barquist L, Vogel J
Accelerating discovery and functional analysis of small RNAs with new technologies
Annu Rev Genet 2015, 49: 367-394
Wheeler NE, Gardner PP, Barquist L
Machine learning identifies signatures of host adaptation in the bacterial pathogen Salmonella enterica
PLos Genetics In press
Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, Page AJ, Langridge GC, Quail MA, Keane JA, Parkhill J
The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries
Bioinformatics 2016, 32(7): 1109-1111
Wheeler NE, Barquist L, Kingsley RA, Gardner PP
A profile-based method for identifying functional divergence of orthologous genes in bacterial genes
Bioinformatics 2016, 32(23): 3566-3574
Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J
Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions
Nature 2016, 529(7587): 496-501