LncRNA and Infection Biology

Our research
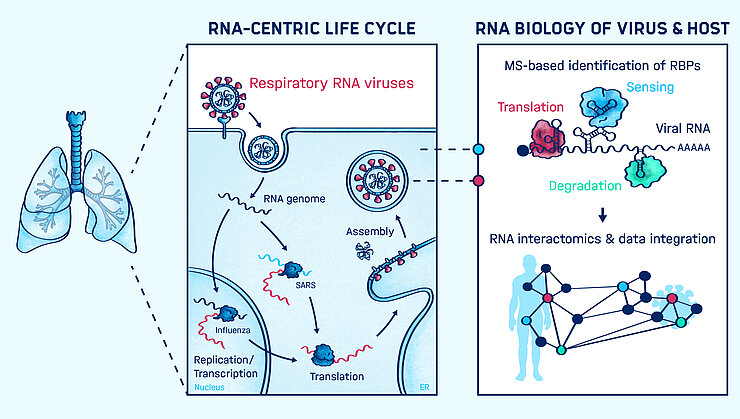
To effectively combat invading pathogens, host cells need to be able to rapidly adjust their gene expression programs and mount an effective host response. In addition to messenger RNA (mRNA), thousands of so called long non-coding RNAs (lncRNAs) are actively transcribed and specifically regulated as a result of bacterial or viral infections. While lncRNA resembles their protein coding counterparts in length, splicing structure, and biochemical properties, they do not serve as templates for protein synthesis. Hence, their physiological functions and biochemical mechanisms are challenging to dissect and in many cases remain poorly understood.
Recent breakthroughs in DNA sequencing technologies led to the realization that many lncRNAs are potent regulators of various gene expression programs, including the host response to pathogens. Knowledge of hundreds to thousands of pathogen-responsive lncRNAs can be considered a treasure trove for discovering novel mechanisms of gene regulation and host defense strategies.
Our group aims to decipher the genetic code controlling lncRNA function by obtaining a quantitative understanding of their molecular interactions and decoding the sequence features or structural elements that mediate these interactions. We seek to elucidate the composition of lncRNA complexes and aim to identify biochemical interactions that enable lncRNA functions. In this context, we are particularly interested in broadly exploring our recent finding that lncRNAs can modulate proteins and control their ability to assemble higher-order ribonucleoprotein complexes. Our group is developing and applying cutting-edge technologies to characterize direct interactions of individual RNA species with proteins at high resolution and in a quantitative manner. Ultimately, we hope to utilize insights into the mechanisms of lncRNA function in order to improve our understanding and ability to treat infectious disease.
Our research
To effectively combat invading pathogens, host cells need to be able to rapidly adjust their gene expression programs and mount an effective host response. In addition to messenger RNA (mRNA), thousands of so called long non-coding RNAs (lncRNAs) are actively transcribed and specifically regulated as a result of bacterial or viral infections. While lncRNA resembles their protein coding counterparts in length, splicing structure, and biochemical properties, they do not serve as templates for protein synthesis. Hence, their physiological functions and biochemical mechanisms are challenging to dissect and in many cases remain poorly understood.
Recent breakthroughs in DNA sequencing technologies led to the realization that many lncRNAs are potent regulators of various gene expression programs, including the host response to pathogens. Knowledge of hundreds to thousands of pathogen-responsive lncRNAs can be considered a treasure trove for discovering novel mechanisms of gene regulation and host defense strategies.
Our group aims to decipher the genetic code controlling lncRNA function by obtaining a quantitative understanding of their molecular interactions and decoding the sequence features or structural elements that mediate these interactions. We seek to elucidate the composition of lncRNA complexes and aim to identify biochemical interactions that enable lncRNA functions. In this context, we are particularly interested in broadly exploring our recent finding that lncRNAs can modulate proteins and control their ability to assemble higher-order ribonucleoprotein complexes. Our group is developing and applying cutting-edge technologies to characterize direct interactions of individual RNA species with proteins at high resolution and in a quantitative manner. Ultimately, we hope to utilize insights into the mechanisms of lncRNA function in order to improve our understanding and ability to treat infectious disease.
Prof Dr Mathias Munschauer
Decoding how long non-coding RNAs work mechanistically will enable us to harness their properties for the treatment of human disease.

Mathias Munschauer studied Biotechnology at the University of Applied Sciences in Mannheim, Germany. Early as an undergraduate student, he joined the lab of Thomas Tuschl at Rockefeller University in New York and started to work with RNA and RNA-binding proteins. He completed his thesis research on the RNA-binding protein FMRP under the supervision of Thomas Tuschl. He then joined an international PhD exchange program jointly operated by the Max-Delbrück Center for Molecular Medicine in Berlin (working with Markus Landthaler) and at New York University (working with Christine Vogel). In the Landthaler lab, Mathias Munschauer pioneered technologies to capture the mRNA-bound proteome and display its global protein occupancy pattern on protein coding transcripts for the first time.
For his postdoctoral research, he joined the lab of Eric Lander at the Broad Institute of MIT and Harvard, where he dissected how long non-coding RNAs function mechanistically and identified a novel RNA-dependent topoisomerase complex. Following his tenure in the Lander lab, he was awarded a Helmholtz Young Investigator Group Leader position and joined HIRI in July 2019 to start his independent research group.
Selected Publications
Munschauer, M.#, Nguyen, C.T., Sirokman, K., Hartigan C. R., Hogstrom, L., Engreitz, J. M, Fulco, C. P., Subramanian V., Chen, J., Ulirch J. C., Schenone, M. Guttman, M., Carr, S.A. Lander, E. S.# (2018). The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature, 561(7721):132-136. doi.org/10.1038/s41586-018-0453-z
Munschauer, M. and Vogel, J. (2018) Nuclear lncRNA stabilization in the host response to bacterial infection. EMBO J, 37(13). pii: e99875. doi.org/10.15252/embj.201899875.
Khajuria R.K., Munschauer, M., Ulirsch J.C., Fiorini C., Ludwig L.S., McFarland S.K., Abdulhay N.J., Specht H., Keshishian H., Mani D.R., Jovanovic M., Ellis S.R., Fulco C.P., Engreitz J.M., Schütz S., Lian J., Gripp K.W., Weinberg O.K., Pinkus G.S., Gehrke L., Regev A., Lander E.S., Gazda H.T., Lee W.Y., Panse V.G., Carr S.A., Sankaran V.G. (2018). Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell, 173(1):90-103.e19. doi.org/10.1016/j.cell.2018.02.036
Fulco, C. P., Munschauer, M., Anyoha, R., Munson, G., Grossman, S. R., Perez, E. M., Kane, M., Cleary B., Lander E. S., Engreitz E. M. (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science, 354(6313):769-773. doi.org/10.1126/science.aag2445
Baltz, A.G.*, Munschauer, M.*, Schwanhaeusser, B., Vasile, A., Murakawa, Y., Schueler, M., Youngs, N., Penfold-Brown, D., Drew, K., Milek, M., Wyler, E., Bonneau, R., Selbach, M., Dieterich, C., and Landthaler, M. (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Molecular Cell 46(5):674-90. doi.org/10.1016/j.molcel.2012.05.021



![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/3/e/csm_Munschauer_Mathias_c_HIRI_Nik_Schoelzel-2_52bc4bb68f.jpg)