Microbial Natural Products

Our Research
Many pathogens are able to generate resistances against drugs like antibiotics leading to an uncontrolled spreading of pathogenic strains. However, an efficient treatment of numerous diseases is missing due to the lack of specific drugs. Therefore, a major task in pharmaceutical research is the identification and development of new agents. Interesting targets are natural products, so-called secondary metabolites, synthesized by microbes, plants and fungi. These agents can, for example, act as antibiotics, anti-cancer drugs, cholesterol-lowering agents, immunosuppressants, anti-parasitics and anti-diabetics.
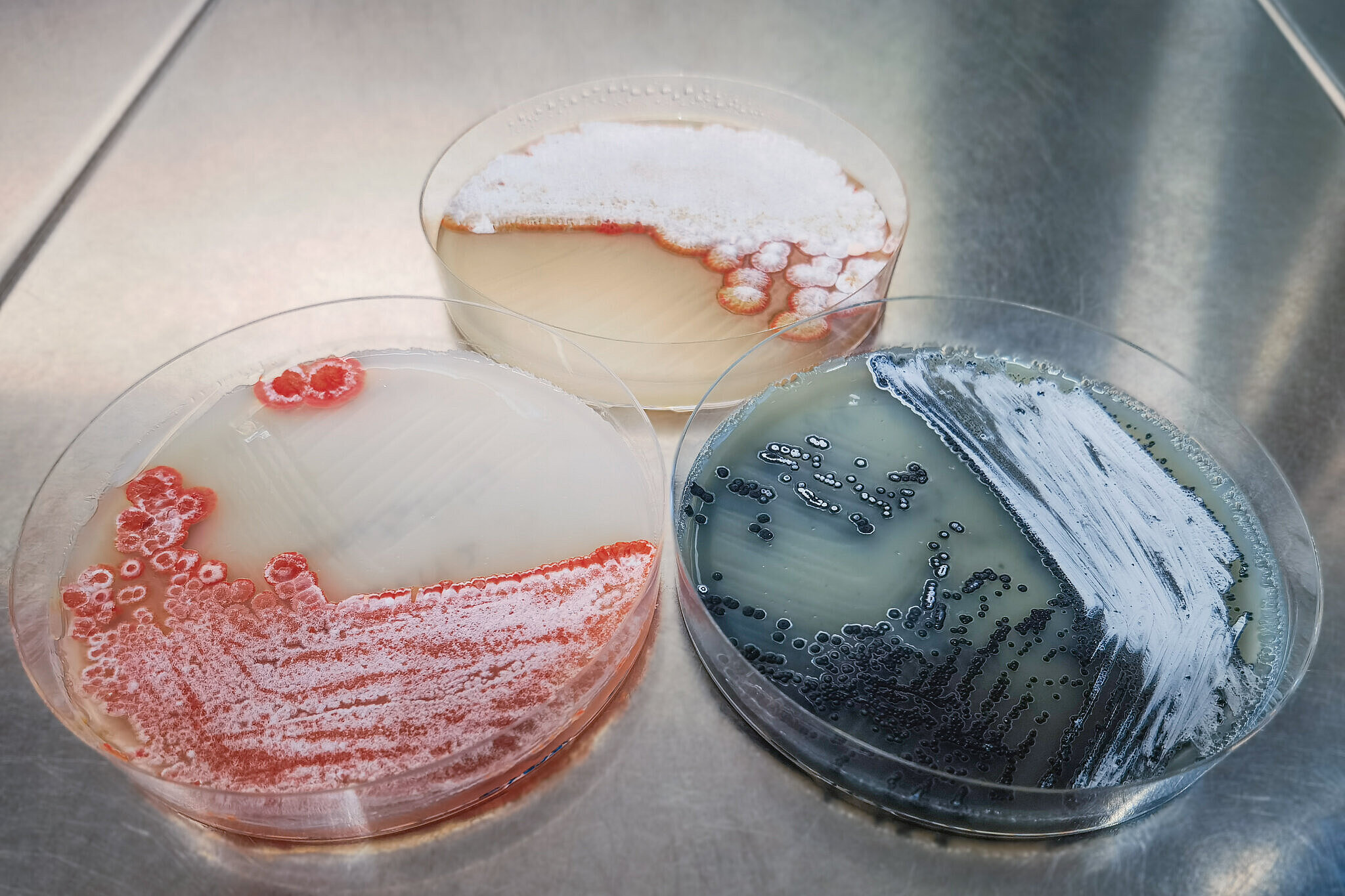
One of the large-scale producers of natural products are myxobacteria, which are living in soil and secrete a number of compounds not only to kill their microbial competitors or enemies. The department “Microbial Natural Products” investigates the chemistry, production, regulation and mode-of-action of secreted metabolites from myxobacteria and, more recently, also from other natural producers such as actinomycetes, a different type of soil-living bacteria.
The scientists study the secondary metabolism in these microorganisms underpinning their efforts with whole-genome sequencing of model strains. Recently, they revealed the largest bacterial genome yet discovered in the myxobacterium Sorangium cellulosum. This bacterium has a huge capacity to produce natural products. The data allow the optimization of metabolite production in these microbes and the expression of complete pathways in heterologous hosts. Furthermore, it is possible to perform predictable alterations to the agents’ structures by genetic engineering. Experiments are underway to “mine” the sequenced genomes for novel compounds using state-of-the-art analytical techniques as well as to deepen the understanding of host microbiology.
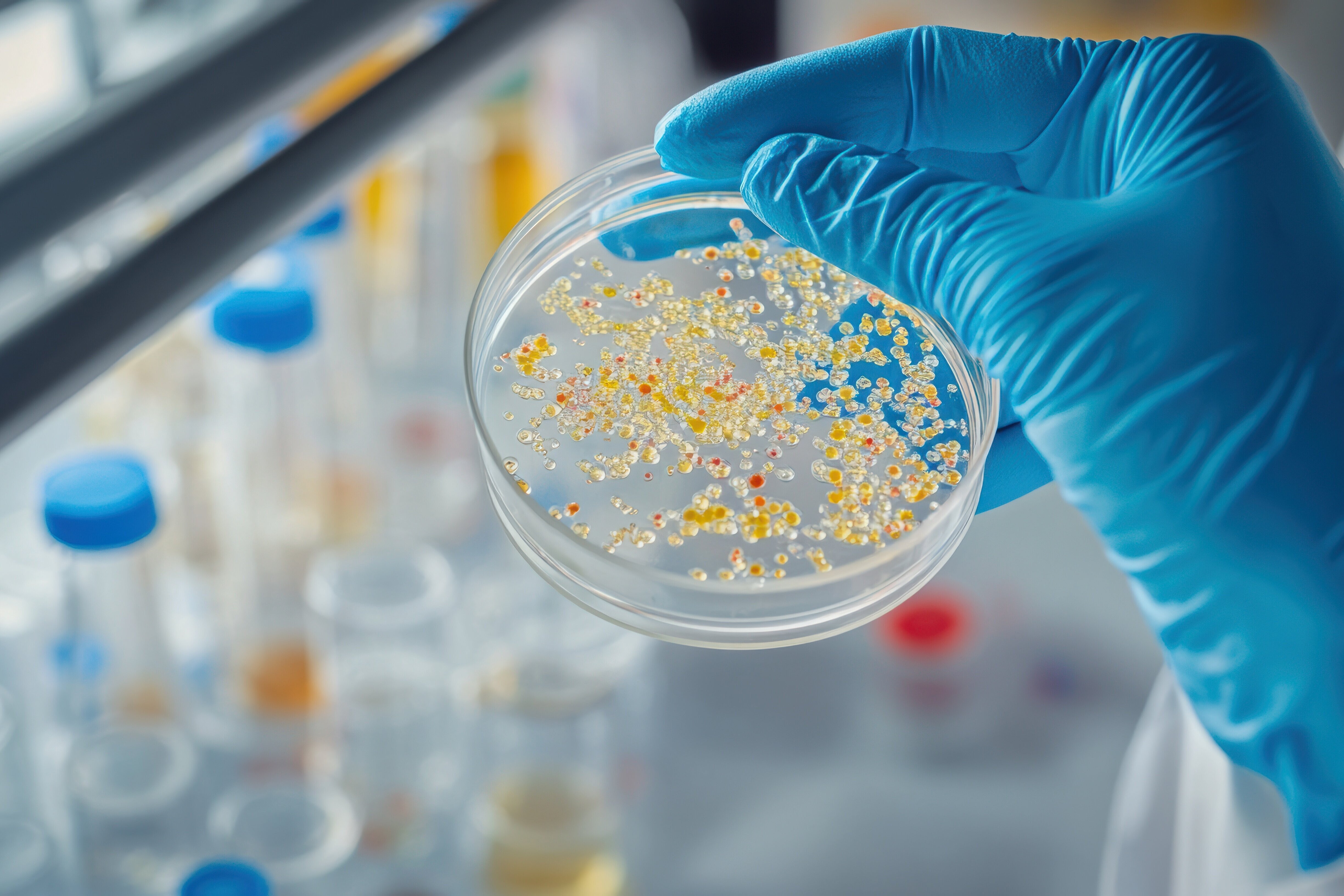
The scientists also run a world-wide myxobacterial strain-discovery program, which results in the identification of new myxobacterial species, genera and families. Once a new strain has been successfully adapted to growth under laboratory conditions, we use state-of-the-art mass spectrometric techniques to screen the strain's metabolite profile with respect to the presence of known myxobacterial secondary metabolites. At the same time we perform a range of biological assays, using indicator organisms as well as cell-based screening approaches to discover novel compounds exhibiting a potentially interesting activity. Following optimization of the production of candidate compounds, cultivation of the producing strain is upscaled and the target compounds are purified using liquid chromatography methods. Thereafter, structure elucidation using a panel of analytical techniques including multidimensional NMR spectroscopy is carried out. In addition, further cell-based experiments are performed in order to investigate in detail the mode-of-action of a novel natural product.
Scientists in the interdisciplinary research group combine a broad spectrum of techniques including microbiological, molecular-biological, genetic, biochemical, analytical and bioengineering methods.
Our Research
Many pathogens are able to generate resistances against drugs like antibiotics leading to an uncontrolled spreading of pathogenic strains. However, an efficient treatment of numerous diseases is missing due to the lack of specific drugs. Therefore, a major task in pharmaceutical research is the identification and development of new agents. Interesting targets are natural products, so-called secondary metabolites, synthesized by microbes, plants and fungi. These agents can, for example, act as antibiotics, anti-cancer drugs, cholesterol-lowering agents, immunosuppressants, anti-parasitics and anti-diabetics.
One of the large-scale producers of natural products are myxobacteria, which are living in soil and secrete a number of compounds not only to kill their microbial competitors or enemies. The department “Microbial Natural Products” investigates the chemistry, production, regulation and mode-of-action of secreted metabolites from myxobacteria and, more recently, also from other natural producers such as actinomycetes, a different type of soil-living bacteria.
The scientists study the secondary metabolism in these microorganisms underpinning their efforts with whole-genome sequencing of model strains. Recently, they revealed the largest bacterial genome yet discovered in the myxobacterium Sorangium cellulosum. This bacterium has a huge capacity to produce natural products. The data allow the optimization of metabolite production in these microbes and the expression of complete pathways in heterologous hosts. Furthermore, it is possible to perform predictable alterations to the agents’ structures by genetic engineering. Experiments are underway to “mine” the sequenced genomes for novel compounds using state-of-the-art analytical techniques as well as to deepen the understanding of host microbiology.
The scientists also run a world-wide myxobacterial strain-discovery program, which results in the identification of new myxobacterial species, genera and families. Once a new strain has been successfully adapted to growth under laboratory conditions, we use state-of-the-art mass spectrometric techniques to screen the strain's metabolite profile with respect to the presence of known myxobacterial secondary metabolites. At the same time we perform a range of biological assays, using indicator organisms as well as cell-based screening approaches to discover novel compounds exhibiting a potentially interesting activity. Following optimization of the production of candidate compounds, cultivation of the producing strain is upscaled and the target compounds are purified using liquid chromatography methods. Thereafter, structure elucidation using a panel of analytical techniques including multidimensional NMR spectroscopy is carried out. In addition, further cell-based experiments are performed in order to investigate in detail the mode-of-action of a novel natural product.
Scientists in the interdisciplinary research group combine a broad spectrum of techniques including microbiological, molecular-biological, genetic, biochemical, analytical and bioengineering methods.
Prof Dr Rolf Müller
We mainly focus on the discovery of microbial natural products in order to develop new antibiotics to combat infectious diseases.

Rolf Müller studied pharmacy at Bonn University and obtained his PhD at the Department of Pharmaceutical Biology. In 1996, he went to the Department of Chemistry at the University of Washington in Seattle, USA . This was when he began to investigate the production of antibiotics in bacteria. Two years later he returned to Germany as a junior group leader at the German Research Centre for Biotechnology (GBF, now the HZI) in Braunschweig. In 2000, he completed his habilitation thesis at the Technische Universität Braunschweig on the biosynthesis of antibiotics in actinomycetes and myxobacteria.
Since October 2003, Rolf Müller has held a chair as professor of Pharmaceutical Biotechnology at Saarland University. In 2009 Rolf Müller became the managing director of the Helmholtz Institute for Pharmaceutical Research Saarland (HIPS), heads the Department of “Microbial Natural Products” (MINS) and co-founded the PharmBioTec GmbH in Saarbrücken. Within the German Center for Infection Research (DZIF) Rolf Müller coordinates the thematic unit “Novel Antibiotics.”
His research has already earned the Phoenix-Pharmacy Research Award three times (2001,2007,2016) the DECHEMA Award for the Natural Products Research (2002), the BioFuture Award of the GermanFederal Ministry for Education and Research (2003), and the DECHEMA Award of the Max-Buchner Research Foundation (2010).
Since 2012 he has been a member of the German National Academy of Science and Engineering. In 2016 he also become elected member of National Academy of Sciences (Leopoldina). Additionally, he has been awarded the Gottfried Wilhelm Leibniz Prize 2021.
Selected Publications
Xie, F., Zhao, H., Liu, J., Yang, X., Neuber, M., Agrawal, A.A., Kaur, A., Herrmann, J., Kalinina, O.V., Wie, X., Müller, R.,* Fu, C.* (2024) Autologous DNA mobilization and multiplication expedite natural products discovery from bacteria. Science, 386 ,eabq7333. DOI:10.1126/science.abq7333
Fu, C., Liu, Y., Walt, C., Rasheed, S., Bader, C., Lukat, P., Neuber, M., Haeckl, F., Blankenfeldt, W., Kalinina, O., and Müller, R.* (2024) Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins. Nat. Commun. 15, 791. DOI: 10.1038/s41467-024-44953-5
Garcia, R., Popoff, A., Bader, C.D., Löhr, J., Walesch, S., Walt, C., Boldt, J., Bunk, B., Haeckl, F.P.J., Gunesch, A.P., Birkelbach, J., Nübel, U., Pietschmann, T., Bach, T., and Müller, R.* (2024) Discovery of the Pendulisporaceae: An extremotolerant myxobacterial family with distinct sporulation behavior and prolific specialized metabolism. Chem. DOI: 10.1016/j.chempr.2024.04.019
Hofer, W., Deschner, F., Jézéquel, G., Pessanha de Carvalho, L., Abdel-Wadood, N., Pätzold, L., Bernecker, S., Morgenstern, B., Kany, A.M., Große, M., Stadler, M., Bischoff, M., Hirsch, A.K.H., Held, J., Herrmann, J.,* and Müller, R.* (2024) Functionalization of Chlorotonils: Dehalogenil as promising lead compound for in vivo application. Angew. Chem. Int. Ed., 63, e202319765. DOI: 10.1002/anie.202319765
Kling, A., Lukat, P., Almeida, D.V., Bauer, A., Fontaine, E, Sordello, S., Zaburannyi, N., Herrmann, J., Wenzel, S.C., König, C., Ammerman, N.C., Barrio, M.B., Borchers, K., Bordon-Pallier, F., Brönstrup, M., Courtemanche, G., Gerlitz, M., Geslin, M., Hammann, P., Heinz, D.W., Hoffmann, H., Klieber, S., Kohlmann, M., Kurz, M., Lair, C., Matter, H., Nuermberger, E., Sandeep T., Fraisse, L., Grosset, J.H., Lagrange, S. and Müller, R.* (2015) Targeting DnaN for tuberculosis therapy using novel griselimycins, Science, 348 (6239): 1106-12. DOI: 10.1126/science.aaa4690
A complete list of publications can be found on the HIPS website.
Technology Offers
The following technologies have been developed and patented by the department Microbial Natural Products:
Novel Myxopyronin Analogues and Myxopyronin Genecluster for Mutasynthesis
Cystobactamides – novel antibacterials against gram-negative pathogens
Highly active Chlorotonil A Derivatives for the treatment of Malaria
Novel Darobactin Derivatives - highly active antibacterials against gram-negative bacteria
Newsroom
Are you interested in a bachelor or master thesis? We are looking forward to your request!

![[Translate to English:] 3D-kontrastierte Oberflächenstruktur von LasB (beige) mit einem Inhibitor der 3. Generation (dunkelblau), der Zink (grau) bindet.](/fileadmin/_processed_/8/b/csm_LasB_Foto_b7734ae7a4.png)