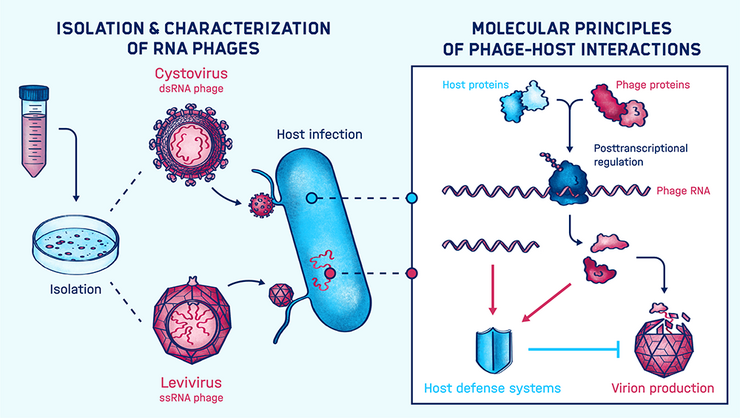
Molecular Principles of RNA Phages

Our Research
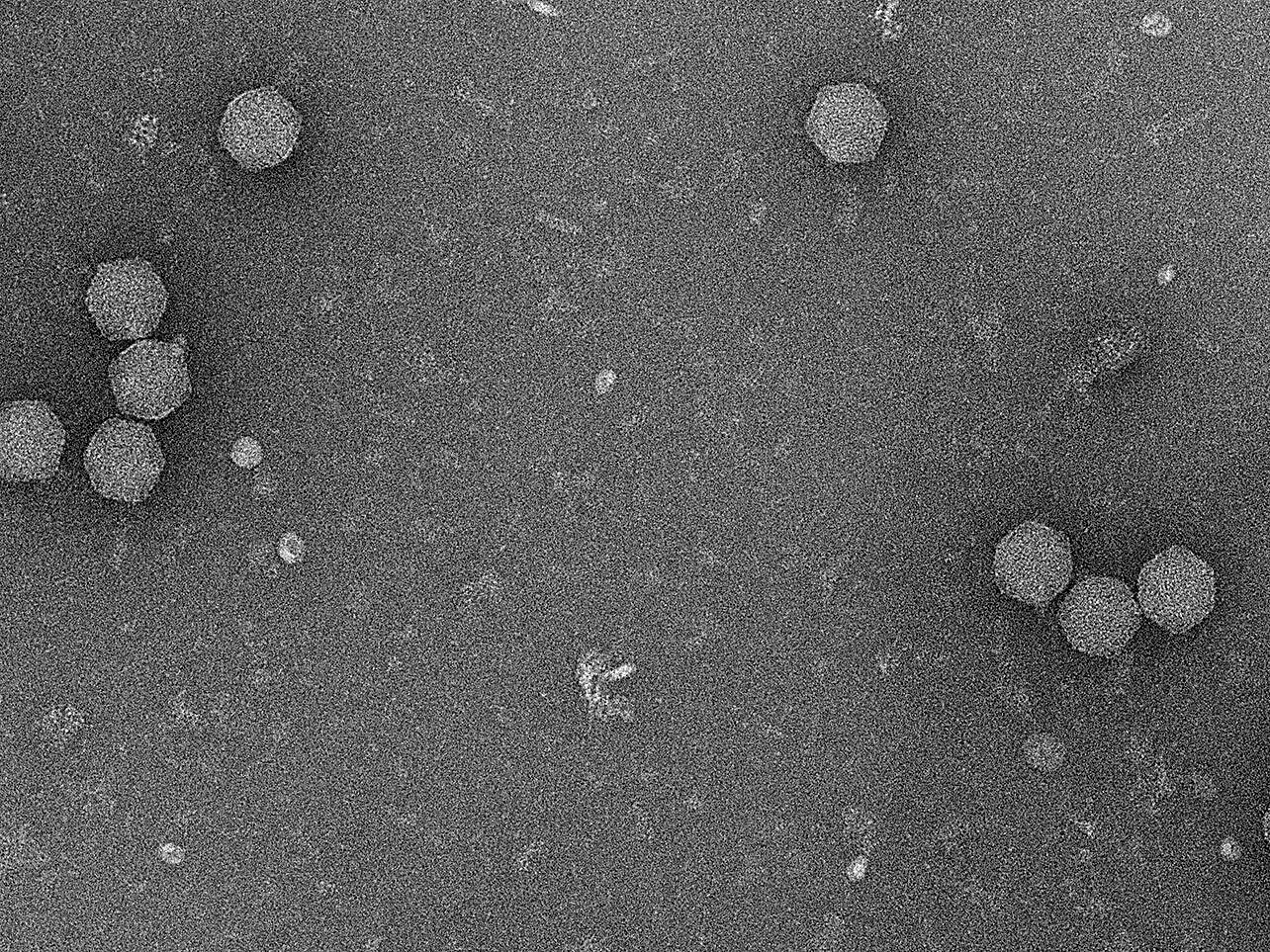
The emerging crisis of antibiotic-resistant bacteria demands the development of novel treatment options for bacterial infections. Phages as the natural predators of bacteria can be exploited to support this process. However, for this, it is crucial to have suitable phages at hand and understand how they function. RNA phages, unlike DNA phages, are heavily understudied. Their characterization will help to unfold the full potential of phages in the design of novel antimicrobial therapies and offers the opportunity to yield new molecular tools and technologies.
Jens Hör’s group works on the molecular principles of RNA phages in the context of phage-host interactions. They seek to understand how RNA phages take over their hosts during infection and how the hosts defend themselves against the invading phages. In addition, they isolate and functionally characterize new RNA phages.
The group employs classical biochemistry and molecular biology methods as well as state-of-the-art high-throughput methods to analyze the unique lifestyle of RNA phages in concert with their host. Their overall goal is to advance our understanding of RNA phages and how they can be exploited to develop novel antibacterial strategies.
Our Research
The emerging crisis of antibiotic-resistant bacteria demands the development of novel treatment options for bacterial infections. Phages as the natural predators of bacteria can be exploited to support this process. However, for this, it is crucial to have suitable phages at hand and understand how they function. RNA phages, unlike DNA phages, are heavily understudied. Their characterization will help to unfold the full potential of phages in the design of novel antimicrobial therapies and offers the opportunity to yield new molecular tools and technologies.
Jens Hör’s group works on the molecular principles of RNA phages in the context of phage-host interactions. They seek to understand how RNA phages take over their hosts during infection and how the hosts defend themselves against the invading phages. In addition, they isolate and functionally characterize new RNA phages.
The group employs classical biochemistry and molecular biology methods as well as state-of-the-art high-throughput methods to analyze the unique lifestyle of RNA phages in concert with their host. Their overall goal is to advance our understanding of RNA phages and how they can be exploited to develop novel antibacterial strategies.
Jun Prof Dr Jens Hör
RNA phages are a heavily understudied group of phages. We seek to better understand their molecular principles and how they can be exploited to develop novel antibacterial strategies.

Jens Hör studied life and medical sciences at the University of Bonn (Germany) and received his PhD from the University of Würzburg (Germany) in 2020. During these studies, he focused on the analysis of the global bacterial RNA-protein complexome. He then joined the Weizmann Institute of Science (Rehovot, Israel) as a postdoc, where he worked on solving the mechanisms of bacterial anti-phage defense systems. Since 2024, he has been a research group leader at the Helmholtz Institute for RNA-based Infection Research (HIRI) and a junior professor at the University of Würzburg.
Selected Publications
Hör J (2025). Advancing RNA phage biology through meta-omics. Nucleic Acids Research 53 (8): gkaf314. DOI: 10.1093/nar/gkaf314
Hör J, Wolf SG, Sorek R (2024). Bacteria conjugate ubiquitin-like proteins to interfere with phage assembly. Nature 631(8022):850-856. DOI: 10.1038/s41586-024-07616-5
Stokar-Avihail A, Fedorenko T, Hör J, Garb J, Leavitt A, Millman A, Shulman G, Wojtania N, Melamed S, Amitai G, Sorek R (2023). Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 186(9):1863-1876.e16. DOI: 10.1016/j.cell.2023.02.029
Hör J, Jung J, Ðurica-Mitić S, Barquist L, Vogel J (2022). INRI-seq enables global cell-free analysis of translation initiation and off-target effects of antisense inhibitors. Nucleic Acids Research 50(22):e128. DOI: 10.1093/nar/gkac838
Hör J, Garriss G, Di Giorgio S, Hack LM, Vanselow JT, Förstner KU, Schlosser A, Henriques-Normark B, Vogel J (2020). Grad-seq in a Gram-positive bacterium reveals exonucleolytic sRNA activation in competence control. The EMBO Journal 39(9):e103852. DOI: 10.15252/embj.2019103852
![[Translate to English:] Rasterelektronenmikroskopische Aufnahme einer Escherichia coli-Zelle (rot), die von Bakteriophagen (grün) infiziert wird.](/fileadmin/_processed_/1/6/csm_Escherichia_coli_phage_c57d3dd91f.jpg)


