Nano Infection Biology

Our working group investigates the cell interaction of respiratory RNA viruses with the help of modern microscopy methods in order to understand how individual viruses communicate with their host cells and how these signals are interpreted by the cells.
The cellular plasma membrane plays a crucial role during virus entry into the cell and also for the assembly of new infectious viruses. It serves the cell on the one hand as a communication interface, but also as a barrier against invading pathogens. Viruses are able to use and modify the plasma membrane, its structure and components in a targeted manner. Our aim is to study these processes at the level of individual viruses and individual proteins.
This nanoscopic view is important because viruses, due to their size and limited genome, have little opportunity to interact with the host cell in order to initiate an infection. The cell's own proteins are bound, an initially rather unspecific process, but then also activated, which in turn appears to occur in a specific manner. We use high- and super-resolution microscopy methods to investigate virus infection on a nanoscopic level in order to understand cellular structures and their dynamics during infection.
Our working group investigates the cell interaction of respiratory RNA viruses with the help of modern microscopy methods in order to understand how individual viruses communicate with their host cells and how these signals are interpreted by the cells.
The cellular plasma membrane plays a crucial role during virus entry into the cell and also for the assembly of new infectious viruses. It serves the cell on the one hand as a communication interface, but also as a barrier against invading pathogens. Viruses are able to use and modify the plasma membrane, its structure and components in a targeted manner. Our aim is to study these processes at the level of individual viruses and individual proteins.
This nanoscopic view is important because viruses, due to their size and limited genome, have little opportunity to interact with the host cell in order to initiate an infection. The cell's own proteins are bound, an initially rather unspecific process, but then also activated, which in turn appears to occur in a specific manner. We use high- and super-resolution microscopy methods to investigate virus infection on a nanoscopic level in order to understand cellular structures and their dynamics during infection.
Dr Christian Sieben
Viruses are nanoscale entities. Over the past decade, we have the reached the technological competence to visualize infection processes on the very scale – an exciting and often surprising view on virus biology.

Christian Sieben studied Biology at the TU Darmstadt with majors in physiology and cell biology. After graduating in plant cell biology, he moved to the Humboldt-Universität zu Berlin to start a PhD in single-cell virology. During this time, he developed microscopy approaches to decipher respiratory virus infection at the level of individual cells. He then moved to EPFL in Switzerland to study super-resolution microscopy techniques, which now allow him to study virus infection at the single protein level. In 2020, Christian started his group Nanoscale Infection Biology (NIBI) at HZI in Braunschweig.
Team








Projects
2025 - 2030 | COMBINE - Identification of Antiviral Targets against Emerging Viruses funded through Horizon Europe. C.S. is PI and coordinator.
2021 - 2025 | Respiratory mucus as a barrier for zoonotic viruses funded through HYI, PhD scholarship from SFB 1449
Since 2022 | Neurotropic influenza A virus infection funded through HYI
Since 2021 | Antiviral drug delivery though respiratory mucus funded through SPRIN-D
Since 2021 | Virus-actin cortex interaction and regulation funded through CSC
Since 2021 | Plasma membrane organization and signalling in virus-cell interaction funded through HYI, CSC, Horizon Europe
2020 - 2024 | Host factors in orthohantavirus-cell entry funded through HYI, HANTadapt (Helmholtz)
Selected Publications
Live-Cell Single-Molecule Imaging of Influenza A Virus-Receptor Interaction
Lukas Broich, Yang Fu, , Fu, Y. and Christian Sieben#
Influenza Virus: Methods and Protocols (2025) DOI: 10.1007/978-1-0716-4326-6_4 #corresponding author
H7N7 viral infection elicits pronounced, sex-specific neuroinflammatory responses in vitro
Lea Gabele, Isabell Bochow, Nele Rieke, Christian Sieben, Kristin Michaelsen-Preusse, Shirin Hosseini, Martin Korte
Frontiers in Cellular Neuroscience (2024) DOI: 10.3389/fncel.2024.1444876
An improved workflow for the quantification of orthohantavirus infection using automated imaging and flow cytometry
Laura Menke, Christian Sieben#
Viruses (2024), DOI: 10.3390/v16020269 #corresponding author
Sieben, C.#, Sezgin, E., Eggeling, C. and Manley, S.#, 2020. Influenza A viruses use multivalent sialic acid clusters for cell binding and receptor activation. PLoS Pathogens, 16(7), p.e1008656. DOI: 10.1371/journal.ppat.1008656
Koehler, M., Delguste, M., Sieben, C., Gillet, L. and Alsteens, D., 2020. Initial Step of Virus Entry: Virion Binding to Cell-Surface Glycans. DOI: 10.1146/annurev-virology-122019-070025



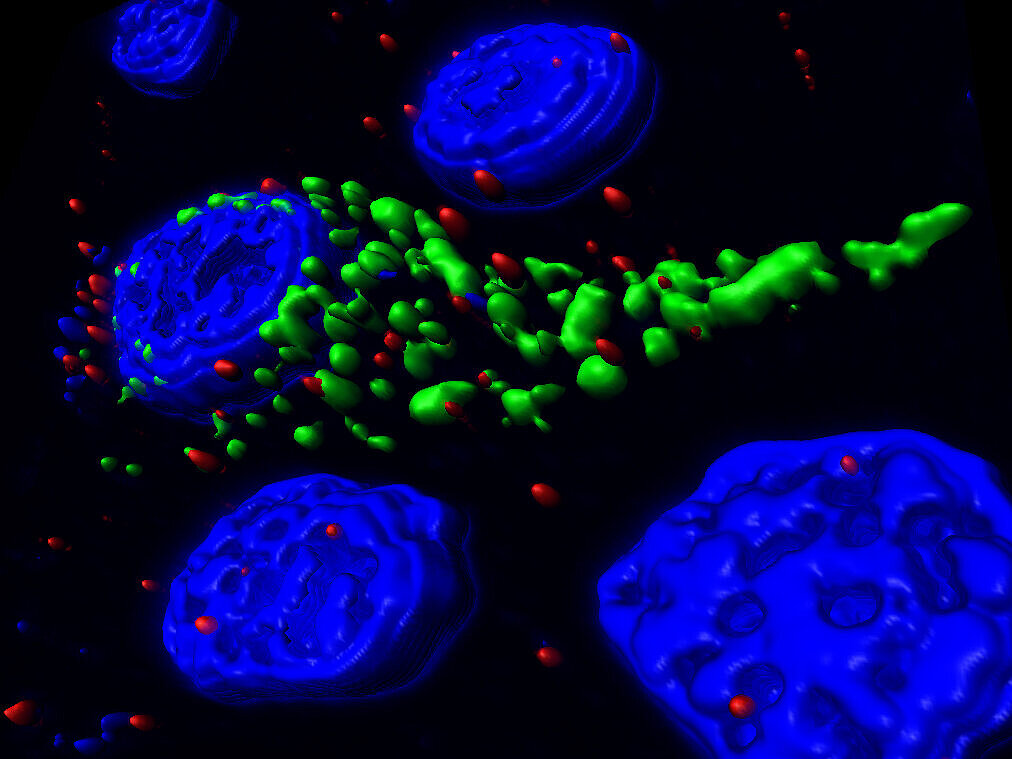
![[Translate to English:] Endogener ASC-Speck, abgebildet in 3D mit dSTORM. © iScience, CC-BY-4.0](/fileadmin/user_upload/HZI/Media_Center/Newsroom/2023/2023_Webnews_LMU_Sieben_c_iScience_CCBY40.png)

