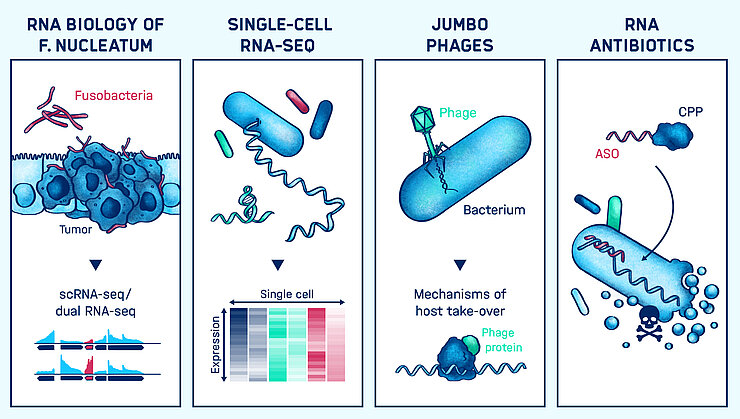
RNA Biology of bacterial infections

Our Research
There are four main highly interactive research areas in the lab:
Discovery of functional RNAs in the microbiome
The major classes of bacterial noncoding RNA are known only for a few species, meaning that the RNA biology of the vast majority of microbes that are important for human health remains unexplored. Exploiting RNA as a programmable antibiotic to treat bacterial infections and dysbiosis requires that we comprehensively understand the major RNA-based pathways and mechanisms in these species. Over the years, we have pioneered a number of RNA deep sequencing-based methods such as differential RNA-seq (Sharma et al. 2010 Nature) or Dual RNA-seq (Westermann et al. 2016 Nature) to understand which and when RNAs are expressed during the course of an infection. Moreover, with Grad-seq (Smirnov et al. 2016 PNAS) we can now classify potentially all cellular transcripts in an unbiased manner based on their biochemical behavior. Using these approaches we are beginning to chart the full cosmos of RNA functions in the human microbiome.
New RNA-binding proteins
Noncoding RNA molecules generally interact with RNA-binding proteins (RBPs) to become functional in the cellular environment. Decades of work in eukaryotes have established RBPs as a pillar of post-transcriptional gene control, with numbers ranging from the hundreds to thousands in vertebrates, invertebrates and yeast cells. Ironically, we know relatively little about the number of RBPs and the mechanisms by which they function in the organisms that seem the easiest to analyse: bacteria. Although bacteria extensively regulate gene expression at the RNA level, our knowledge of global RBPs remains limited to the sRNA chaperone Hfq and the translational repressor CsrA. However, increasing evidence, including the serendipitous discovery of CRISPR-Cas systems, suggests that many RBPs await discovery especially in less well studied microbes.
We have discovered the “osmoregulatory” protein ProQ as a new abundant RBP which binds hundreds cellular RNAs in Salmonella and E. coli (Smirnov et al. 2016 PNAS). Our results for ProQ suggest new molecular mechanisms of bacterial gene control at mRNA 3’ ends and a novel regulatory network whereby ProQ recognizes RNA structure rather than primary sequence (Smirnov et al. 2017 EMBO J Holmqvist et al. 2018 Molecular Cell). We are also addressing the fundamental principles involved in RNA recognition by trying to understand how members of this RBP family use the FinO domain for either highly specific or global RNA recognition in the same cellular environment. Continuing with RBP discovery, we are now exploring the microbiome for new RBPs.
Single-cell RNA biology
The development of single-cell RNA-seq and its ability to describe the gene expression profiles of individual cells rather than the average cellular population is rapidly changing our view of many aspects of biology, including the discovery of new cell types and functions. Single-cell RNA-seq is also emerging as a powerful tool to determine cell-to-cell variability in bacterial pathogenesis, however, technical issues have restricted its use to the analysis of eukaryotic transcriptomes. For example, we have studied how heterogeneous intracellular growth of Salmonella drives the polarization state of individually infected macrophages (Saliba et al. 2017 Nature Microbiology).
Our grand challenge is to fully establish single-cell RNA-seq for bacteria, which is currently hampered by the small amounts of cellular RNA present and the lack of a polyA tail for easy cDNA priming as in eukaryotes. Transcriptomes from single bacteria will provide insight into the basis for phenotypic resistance to antibiotic treatment, stochastic persister formation, and phenotypic diversification for bet hedging, poorly understood phenomena that are threatening our ability to successfully treat bacterial infections. It will also be the next step for developing single-cell Dual RNA-seq to simultaneously study individual host-pathogen interactions. Going beyond simple single-cell transcriptomics, our long-term ambition is to develop robust methodologies for routine high-throughput RNA biochemistry to study post-transcriptional control in single bacterial cells.
The RNA route to manipulation of the microbiota
Our body is colonized by a vast array of bacteria, which together form the human microbiome. The gut alone harbours more than 1,000 bacterial species. An understanding of their individual or synergistic contributions to human health and disease demands a means to interfere and perturb their functions on the species level, with a selectivity that is not possible using currently available antibiotics.
Our ultimate goal is precision manipulation of the microbiota using “programmable antibiotics” in the form of RNA-directed antisense oligonucleotides (ASOs). The same approach can be taken to target antibiotic-resistant pathogens as an alternative to standard antibiotics. These ASOs are coupled to small peptides that carry them inside the bacteria to silence the mRNAs of essential genes. There is already proof-of-principle for this exciting concept, but many open questions remain. What does it take to make these ASOs truly species-specific; how can we minimize potential off-target effects; what are the most effective peptides for delivery; what about the host response and bacterial resistance mechanisms? Since there is unlikely to be a one case-fits-all solution for all microbiome species, this project can only succeed by integrating diverse expertise from RNA biology, microbiology, immunology, single-cell biology, and pharmacy.
Our Research
There are four main highly interactive research areas in the lab:
Discovery of functional RNAs in the microbiome
The major classes of bacterial noncoding RNA are known only for a few species, meaning that the RNA biology of the vast majority of microbes that are important for human health remains unexplored. Exploiting RNA as a programmable antibiotic to treat bacterial infections and dysbiosis requires that we comprehensively understand the major RNA-based pathways and mechanisms in these species. Over the years, we have pioneered a number of RNA deep sequencing-based methods such as differential RNA-seq (Sharma et al. 2010 Nature) or Dual RNA-seq (Westermann et al. 2016 Nature) to understand which and when RNAs are expressed during the course of an infection. Moreover, with Grad-seq (Smirnov et al. 2016 PNAS) we can now classify potentially all cellular transcripts in an unbiased manner based on their biochemical behavior. Using these approaches we are beginning to chart the full cosmos of RNA functions in the human microbiome.
New RNA-binding proteins
Noncoding RNA molecules generally interact with RNA-binding proteins (RBPs) to become functional in the cellular environment. Decades of work in eukaryotes have established RBPs as a pillar of post-transcriptional gene control, with numbers ranging from the hundreds to thousands in vertebrates, invertebrates and yeast cells. Ironically, we know relatively little about the number of RBPs and the mechanisms by which they function in the organisms that seem the easiest to analyse: bacteria. Although bacteria extensively regulate gene expression at the RNA level, our knowledge of global RBPs remains limited to the sRNA chaperone Hfq and the translational repressor CsrA. However, increasing evidence, including the serendipitous discovery of CRISPR-Cas systems, suggests that many RBPs await discovery especially in less well studied microbes.
We have discovered the “osmoregulatory” protein ProQ as a new abundant RBP which binds hundreds cellular RNAs in Salmonella and E. coli (Smirnov et al. 2016 PNAS). Our results for ProQ suggest new molecular mechanisms of bacterial gene control at mRNA 3’ ends and a novel regulatory network whereby ProQ recognizes RNA structure rather than primary sequence (Smirnov et al. 2017 EMBO J Holmqvist et al. 2018 Molecular Cell). We are also addressing the fundamental principles involved in RNA recognition by trying to understand how members of this RBP family use the FinO domain for either highly specific or global RNA recognition in the same cellular environment. Continuing with RBP discovery, we are now exploring the microbiome for new RBPs.
Single-cell RNA biology
The development of single-cell RNA-seq and its ability to describe the gene expression profiles of individual cells rather than the average cellular population is rapidly changing our view of many aspects of biology, including the discovery of new cell types and functions. Single-cell RNA-seq is also emerging as a powerful tool to determine cell-to-cell variability in bacterial pathogenesis, however, technical issues have restricted its use to the analysis of eukaryotic transcriptomes. For example, we have studied how heterogeneous intracellular growth of Salmonella drives the polarization state of individually infected macrophages (Saliba et al. 2017 Nature Microbiology).
Our grand challenge is to fully establish single-cell RNA-seq for bacteria, which is currently hampered by the small amounts of cellular RNA present and the lack of a polyA tail for easy cDNA priming as in eukaryotes. Transcriptomes from single bacteria will provide insight into the basis for phenotypic resistance to antibiotic treatment, stochastic persister formation, and phenotypic diversification for bet hedging, poorly understood phenomena that are threatening our ability to successfully treat bacterial infections. It will also be the next step for developing single-cell Dual RNA-seq to simultaneously study individual host-pathogen interactions. Going beyond simple single-cell transcriptomics, our long-term ambition is to develop robust methodologies for routine high-throughput RNA biochemistry to study post-transcriptional control in single bacterial cells.
The RNA route to manipulation of the microbiota
Our body is colonized by a vast array of bacteria, which together form the human microbiome. The gut alone harbours more than 1,000 bacterial species. An understanding of their individual or synergistic contributions to human health and disease demands a means to interfere and perturb their functions on the species level, with a selectivity that is not possible using currently available antibiotics.
Our ultimate goal is precision manipulation of the microbiota using “programmable antibiotics” in the form of RNA-directed antisense oligonucleotides (ASOs). The same approach can be taken to target antibiotic-resistant pathogens as an alternative to standard antibiotics. These ASOs are coupled to small peptides that carry them inside the bacteria to silence the mRNAs of essential genes. There is already proof-of-principle for this exciting concept, but many open questions remain. What does it take to make these ASOs truly species-specific; how can we minimize potential off-target effects; what are the most effective peptides for delivery; what about the host response and bacterial resistance mechanisms? Since there is unlikely to be a one case-fits-all solution for all microbiome species, this project can only succeed by integrating diverse expertise from RNA biology, microbiology, immunology, single-cell biology, and pharmacy.
Prof Jörg Vogel
Until recently, the significance of RNA molecules in infection processes has been greatly underestimated. Today, we know that RNAs interact with a diverse array of molecules of the host cell and the pathogen. To exploit these opportunities, HIRI will pioneer an integrated concept using cutting-edge technologies.

Prof. Jörg Vogel studied biochemistry at Humboldt University, Berlin, Germany, and at Imperial College, London, UK. In 1999, he received his PhD from Humboldt University with a thesis on Group II intron splicing. He spent his postdoctoral years at Uppsala University, Sweden (2000-2001) and as an EMBO fellow at the Hebrew University, Jerusalem, Israel (2002-2003) before he started an Independent Junior Research Group at the Max Planck Institute for Infection Biology (204-2010). He became Full Professor and Director of the Institute for Molecular Infection Biology at the University of Würzburg in 2009. As of 2017, he is the Founding Director of the Helmholtz Institute for RNA-based Infection Research in Würzburg.
Jörg Vogel is an elected member of EMBO, the German National Academy of Sciences, as well as the European and American Academies of Microbiology. In 2017, he received a Leibniz Prize, Germany’s most prestigious research award, for his work combining RNA biology and infection research.
Selected Publications
Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiß S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, Stadler PF, Vogel J (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464 250-255
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602-7
Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J (2013) Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153: 426–437
Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J (2016) Grad-seq guides the discovery of ProQ as a major small RNA binding protein. PNAS 113: 11591-6
Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J (2016) Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529: 496-501
Technology Offers
The following technologies have been developed and patented by the department "RNA Biology of bacterial infections":
Novel ASO-Based mRNA Silencing Technology for Phage-Host Interaction Research