RNA Biology of gram-positive bacteria

Our Research
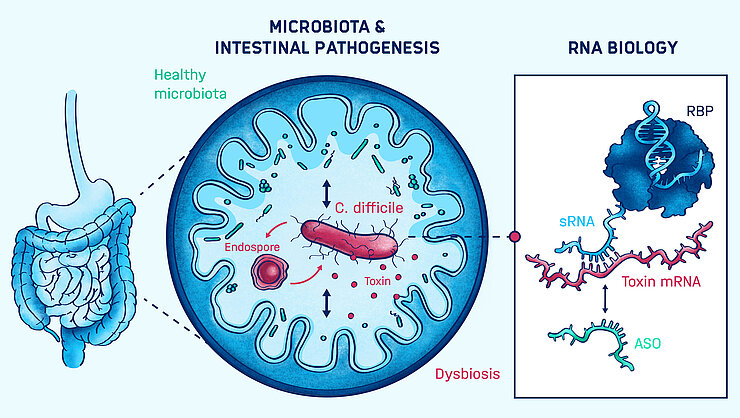
Enteric infections and their clinical outcome are often associated with underlying microbial dysbiosis, an imbalance in the microbiota. Clostridioides difficile—a leading cause of antibiotic-associated diarrhea—is a prime example: In healthy individuals, infection with C. difficile results in asymptomatic colonization. However, after antibiotic treatment, the enteric pathogen causes a wide spectrum of diseases. Therefore, a mechanistic understanding of virulence regulation in the context of pathogen-commensal interactions is paramount for the development of novel RNA-based therapeutics.
Franziska Faber’s group addresses this goal through the molecular characterization of virulence regulation, with a focus on post-transcriptional regulation by non-coding RNAs and their cognate RNA-binding proteins. They also identify the microbial and host signals that regulate the virulence gene expression. Furthermore, the Faber group establishes antisense oligomer-based targeting of genes and pathways essential for virulence.
Their work combines technologies from the fields of RNA biology, biochemistry, metabolomics, genetics and microbiology. The overarching goal of the Faber group is to identify and functionally characterize novel RNA-based mechanisms of virulence regulation that can be exploited as therapeutic targets.
Our Research
Enteric infections and their clinical outcome are often associated with underlying microbial dysbiosis, an imbalance in the microbiota. Clostridioides difficile—a leading cause of antibiotic-associated diarrhea—is a prime example: In healthy individuals, infection with C. difficile results in asymptomatic colonization. However, after antibiotic treatment, the enteric pathogen causes a wide spectrum of diseases. Therefore, a mechanistic understanding of virulence regulation in the context of pathogen-commensal interactions is paramount for the development of novel RNA-based therapeutics.
Franziska Faber’s group addresses this goal through the molecular characterization of virulence regulation, with a focus on post-transcriptional regulation by non-coding RNAs and their cognate RNA-binding proteins. They also identify the microbial and host signals that regulate the virulence gene expression. Furthermore, the Faber group establishes antisense oligomer-based targeting of genes and pathways essential for virulence.
Their work combines technologies from the fields of RNA biology, biochemistry, metabolomics, genetics and microbiology. The overarching goal of the Faber group is to identify and functionally characterize novel RNA-based mechanisms of virulence regulation that can be exploited as therapeutic targets.
RNAs have emerged as important regulators of virulence in C. difficile. We seek to characterize the molecular mechanisms of these regulators and how they can be leveraged for the treatment of C. difficile infections.
Prof. Dr. Franziska Faber

Franziska Faber studied biology at the Martin-Luther University (Halle/Saale, Germany). Her doctoral work at the Robert Koch Institute (Wernigerode, Germany) introduced her to the field of enteric bacterial infections. In 2011, she started her postdoctoral work in the lab of Andreas Bäumler at the University of California (Davis, USA). Here, she studied the mechanisms by which antibiotic treatment lowers “colonization resistance” against the enteric pathogen Salmonella enterica. More specifically, she explored how inflammation-associated changes in intestinal nutrients contribute to intestinal colonization of S. enterica. In 2018, she joined the Center for Infectious Disease Research (ZINF) in Würzburg as a Young Investigator to study the previously unexplored RNA biology of the intestinal pathogen Clostridioides difficile. Since 2021, she has been a Junior Professor at the Institute of Molecular Infection Biology (IMIB) and has since headed the ROSE group at Julius-Maximilians-Universität (JMU) Würzburg, which is associated with the HIRI. In 2023, she was appointed to a W2 professorship at the Institute for Hygiene and Microbiology at JMU.
Selected Publications
Lenče T*, Sulzer J*, Andress K, Gribling-Burrer A-S, Lamm-Schmidt V, Barquist L, Smyth RP, Faber F (2024)
The conserved noncoding RNA ModT coordinates growth and virulence in Clostridioides difficile.
PLoS Biology 22(12):e3002948
Fuchs M, Lamm-Schmidt V, Lenče T, Sulzer J, Bublitz A, Wackenreuter J, Gerovac M, Strowig T, Faber F (2023)
A network of small RNAs regulates sporulation initiation in C. difficile
EMBO Journal 42(12):e112858
Fuchs M*, Lamm-Schmidt V*, Sulzer J, Ponath F, Jenniches L, Kirk JA, Fagan RP, Barquist L, Vogel J, Faber F (2021)
An RNA-centric global view of the clinically important bacterium Clostridioides difficile reveals broad activity of Hfq in a Gram-positive species
PNAS 118(25):e2103579118
Lamm-Schmidt V, Fuchs M, Sulzer J, Gerovac M, Hör J, Dersch P, Vogel J, Faber F (2021)
Grad-seq identifies KhpB as a global RNA-binding protein in Clostridioides difficile that regulates toxin production
microLife 2:uqab004
Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, Bäumler AJ (2016)
Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion
Nature 534(7609):697-699