Author count: 15
Gupta D., Patinios C., Bassett H.V., Kibe A., ..., Kamm C., Wang Y., ..., Toussaint C., Calvin I., ..., Migur A., Schut F., ... , ..., Beisel C.L.
(2025)
Targeted DNA ADP-ribosylation triggers templated repair in bacteria and base mutagenesis in eukaryotes
Nat.Biotechnol.
Author count: 6
Eggenschwiler R., Hoffmann T., Dmytrenko O., ..., Beisel C.L., Cantz T.
(2025)
PAM-interacting domain turn-helix 51 motifs can improve Cas9-SpRY activity
Nucleic Acid Research,
53
(15)
Author count: 5
Arifah A.Q., Vento J.M., Kurrer I., Achmedov T., Beisel C.L.
(2025)
Cas9-independent tracrRNA cytotoxicity in Lacticaseibacillus paracasei
microLife,
6
(uqaf013)
Author count: 7
Westarp P., Keller T., Brand J., Horvat S., Albrecht K., ... , Beisel C., Groll J.
(2025)
Efficient encapsulation of CRISPR-Cas9 RNP in bioreducible nanogels and release in a cytosol-mimicking environment
Discov.Nano,
20
(1)
Author count: 7
Feussner M., Migur A., Mitrofanov A., Alkhnbashi O.S., Backofen R., ... , Beisel C.L., Weinberg Z.
(2025)
Disparate mechanisms counteract extraneous CRISPR RNA production in type II-C CRISPR-Cas systems
microLife,
6
Author count: 13
Della Valle S., Orsi E., Creutzburg S.C.A., Jansen L.F.M., Pentari E.N., Beisel C.L., Steel H., Nikel P.I., Staals R.H.J., Claassens N.J., van der Oost J., ... , Huang W.E., Patinios C.
(2025)
Streamlined and efficient genome editing in Cupriavidus necator H16 using an optimised SIBR-Cas system
Trends Biotechnol.
Author count: 9
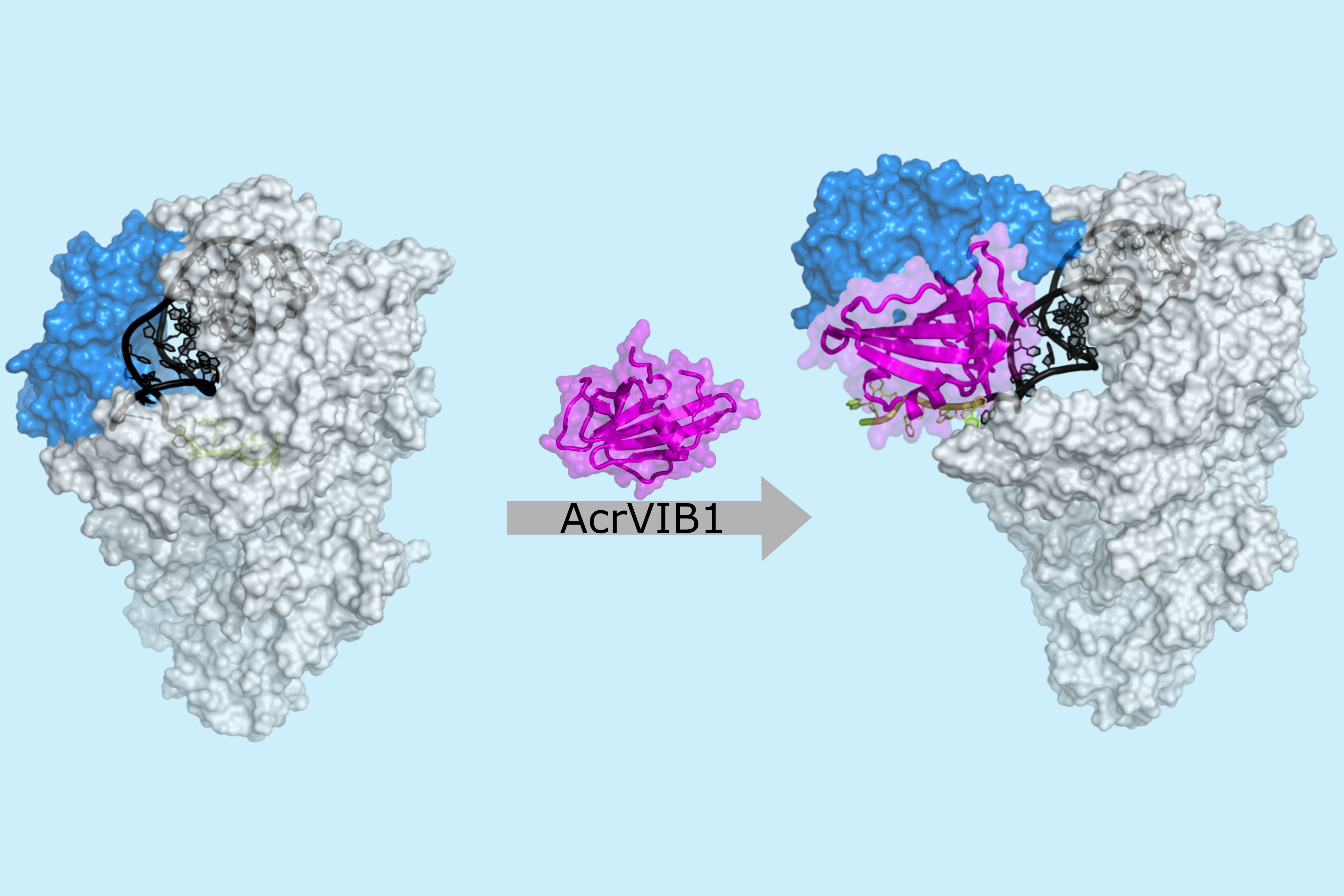
Wandera K.G., Schmelz S., Migur A., Kibe A., Lukat P., Achmedov T., Caliskan N., ... , Blankenfeldt W., Beisel C.L.
(2025)
AcrVIB1 inhibits CRISPR-Cas13b immunity by promoting unproductive crRNA binding accessible to RNase attack
Mol.Cell,
85
(6)
Author count: 5
Schut F.T., Hallmark T., Dmytrenko O., Jackson R.N., Beisel C.L.
(2025)
Purification and in vivo, cell-free, and in vitro characterization of CRISPR-Cas12a2
(7)
Author count: 11
Kibe A., Buck S., Gribling-Burrer A.S., Gilmer O., Bohn P., Koch T., Mireisz C.N., Schlosser A., Erhard F., ... , Smyth R.P., Caliskan N.
(2025)
The translational landscape of HIV-1 infected cells reveals key gene regulatory principles
Nat Struct.Mol.Biol.
Author count: 10
Jabalera Y., Tascon I., Samperio S., Lopez-Alonso J.P., Gonzalez-Lopez M., Aransay A.M., Abascal-Palacios G., Beisel C.L., ... , Ubarretxena-Belandia I., Perez-Jimenez R.
(2024)
A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection
Nat.Biotechnol.
Author count: 2
Kamm C., Beisel C.L.
(2024)
Phages to the rescue: in situ editing of the gut microbiota
Trends Microbiol.,
32
(10)
Author count: 14
Haynes K.A., Andrews L.B., Beisel C.L., Chappell J., Cuba Samaniego C.E., Dueber J.E., Dunlop M.J., Franco E., Lucks J.B., Noireaux V., Savage D.F., Silver P.A., ... , Smanski M., Young E.
(2024)
Ten Years of the Synthetic Biology Summer Course at Cold Spring Harbor Laboratory
ACS Synth Biol,
13
(9)
Author count: 12
Arias-Rojas A., Arifah A.Q., Angelidou G., Alshaar B., Schombel U., Forest E., Frahm D., Brinkmann V., Paczia N., Beisel C.L., ... , Gisch N., Iatsenko I.
(2024)
MprF-mediated immune evasion is necessary for Lactiplantibacillus plantarum resilience in the Drosophila gut during inflammation
PLoS Pathogens,
20
(8)
Author count: 7
Khakimzhan A., Izri Z., Thompson S., Dmytrenko O., Fischer P., ... , Beisel C., Noireaux V.
(2024)
Cell-free expression with a quartz crystal microbalance enables rapid, dynamic, and label-free characterization of membrane-interacting proteins
Communications biology,
7
(1)
Author count: 17
Birkholz N., Kamata K., Feussner M., Wilkinson M.E., Cuba Samaniego C., Migur A., Kimanius D., Ceelen M., Went S.C., Usher B., Blower T.R., Brown C.M., Beisel C.L., Weinberg Z., Fagerlund R.D., ... , Jackson S.A., Fineran P.C.
(2024)
Phage anti-CRISPR control by an RNA- and DNA-binding helix-turn-helix protein
Nature,
631
(8021)
Author count: 9
Jiao C., Peeck N.L., Yu J., Ghaem Maghami M., Kono S., Collias D., Martinez Diaz S.L., ... , Larose R., Beisel C.L.
(2024)
TracrRNA reprogramming enables direct PAM-independent detection of RNA with diverse DNA-targeting Cas12 nucleases
Nature Communications,
15
(1)
Author count: 12
Vento J.M., Durmusoglu D., Li T., Patinios C., Sullivan S., Ttofali F., van Schaik J., Yu Y., Wang Y., Barquist L., ... , Crook N., Beisel C.L.
(2024)
A cell-free transcription-translation pipeline for recreating methylation patterns boosts DNA transformation in bacteria
Mol Cell
Author count: 24
Benz F., Camara-Wilpert S., Russel J., Wandera K.G., Cepaite R., Ares-Arroyo M., Gomes-Filho J.V., Englert F., Kuehn J.A., Gloor S., Mestre M.R., Cuenod A., Aguila-Sans M., Maccario L., Egli A., Randau L., Pausch P., Rocha E.P.C., Beisel C.L., Madsen J.S., Bikard D., Hall A.R., ... , Sorensen S.J., Pinilla-Redondo R.
(2024)
Type IV-A3 CRISPR-Cas systems drive inter-plasmid conflicts by acquiring spacers in trans
Cell Host.Microbe
Author count: 15
Vialetto E., Miele S., Goren M.G., Yu J., Yu Y., Collias D., Beamud B., Osbelt L., Lourenco M., Strowig T., Brisse S., Barquist L., Qimron U., ... , Bikard D., Beisel C.L.
(2024)
Systematic interrogation of CRISPR antimicrobials in Klebsiella pneumoniae reveals nuclease-, guide-and strain-dependent features influencing antimicrobial activity
Nucleic Acids Res.
Author count: 6
Gawlitt S., Collins S.P., Yu Y., Blackman S.A., Barquist L., Beisel C.L.
(2024)
Expanding the flexibility of base editing for high-throughput genetic screens in bacteria
Nucleic Acids Res.
Author count: 5
Prezza G., Liao C., Reichardt S., Beisel C.L., Westermann A.J.
(2024)
CRISPR-based screening of small RNA modulators of bile susceptibility in Bacteroides thetaiotaomicron
Proceedings of the National Academy of Sciences of the United States of America,
121
(6)
Author count: 8
Almasi E.D.H., Knischewski N., Osbelt L., Muthukumarasamy U., el Mouali Y., Vialetto E., ... , Beisel C.L., Strowig T.
(2024)
An adapted method for Cas9-mediated editing reveals the species-specific role of b
J.Bacteriol.
Author count: 3
Krink N., Nikel P.I., Beisel C.L.
(2024)
A Hitchhiker's guide to CRISPR editing tools in bacteria : CRISPR can help unlock the bacterial world, but technical and regulatory barriers persist
EMBO Rep.
Author count: 7
Yu Y., Gawlitt S., de Andrade e Sousa , Merdivan E., Piraud M., ... , Beisel C.L., Barquist L.
(2024)
Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration
Genome Biology,
25
(1)
Author count: 7
Wimmer F., Englert F., Wandera K.G., Alkhnbashi O.S., Collins S.P., ... , Backofen R., Beisel C.L.
(2024)
Interrogating two extensively self-targeting Type I CRISPR-Cas systems in Xanthomonas albilineans reveals distinct anti-CRISPR proteins that block DNA degradation
Nucleic Acids Res.
Author count: 31
Schmidt N., Ganskih S., Wei Y., Gabel A., Zielinski S., Keshishian H., Lareau C.A., Zimmermann L., Makroczyova J., Pearce C., Krey K., Hennig T., Stegmaier S., Moyon L., Horlacher M., Werner S., Aydin J., Olguin-Nava M., Potabattula R., Kibe A., Dölken L., Smyth R.P., Caliskan N., Marsico A., Krempl C., Bodem J., Pichlmair A., Carr S.A., Chlanda P., ... , Erhard F., Munschauer M.
(2023)
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9
Cell,
186
(22)
Author count: 4
Gawlitt S., Liao C., Achmedov T., Beisel C.L.
(2023)
Shortened CRISPR-Cas9 arrays enable multiplexed gene targeting in bacteria from a smaller DNA footprint
RNA Biol.,
20
(1)
Author count: 2
Patinios C., Beisel C.L.
(2023)
For the CRISPR Fan(zor)atics: RNA-guided DNA endonucleases discovered in eukaryotes
Mol.Cell,
83
(17)
Author count: 26
Esser S.P., Rahlff J., Zhao W., Predl M., Plewka J., Sures K., Wimmer F., Lee J., Adam P.S., McGonigle J., Turzynski V., Banas I., Schwank K., Krupovic M., Bornemann T.L.V., Figueroa-Gonzalez P.A., Jarett J., Rattei T., Amano Y., Blaby I.K., Cheng J.F., Brazelton W.J., Beisel C.L., Woyke T., ... , Zhang Y., Probst A.J.
(2023)
A predicted CRISPR-mediated symbiosis between uncultivated archaea
Nat.Microbiol.
Author count: 5
Krink N., Löchner A.C., Cooper H., Beisel C.L., Di Ventura B.
(2022)
Synthetic biology landscape and community in Germany
Biotechnol.Notes,
3
Author count: 13
Cetin R., Wegner M., Luwisch L., Saud S., Achmedov T., Süsser S., Vera-Guapi A., Müller K., Matthess Y., Quandt E., Schaubeck S., ... , Beisel C.L., Kaulich M.
(2023)
Optimized metrics for orthogonal combinatorial CRISPR screens
Sci.Rep.,
13
(1)
Author count: 9
Collias D., Vialetto E., Yu J., Co K., Almasi E., Rüttiger A.S., Achmedov T., ... , Strowig T., Beisel C.L.
(2023)
Systematically attenuating DNA targeting enables CRISPR-driven editing in bacteria
Nat.Commun.,
14
(1)
Author count: 9
Jiao C., Reckstadt C., König F., Homberger C., Yu J., Vogel J., Westermann A.J., ... , Sharma C.M., Beisel C.L.
(2023)
RNA recording in single bacterial cells using reprogrammed tracrRNAs
Nat.Biotechnol.
Author count: 6
Bravo J.P.K., Hallmark T., Naegle B., Beisel C.L., Jackson R.N., Taylor D.W.
(2023)
RNA targeting unleashes indiscriminate nuclease activity of CRISPR-Cas12a2
Nature,
613
(7944)
Author count: 16
Dmytrenko O., Neumann G.C., Hallmark T., Keiser D.J., Crowley V.M., Vialetto E., Mougiakos I., Wandera K.G., Domgaard H., Weber J., Gaudin T., Metcalf J., Gray B.N., Begemann M.B., ... , Jackson R.N., Beisel C.L.
(2023)
Cas12a2 elicits abortive infection through RNA-triggered destruction of dsDNA
Nature,
613
(7944)
Author count: 19
Wu W.Y., Mohanraju P., Liao C., Adiego-Pérez B., Creutzburg S.C.A., Makarova K.S., Keessen K., Lindeboom T.A., Khan T.S., Prinsen S., Joosten R., Yan W.X., Migur A., Laffeber C., Scott D.A., Lebbink J.H.G., Koonin E.V., ... , Beisel C.L., van der Oost J.
(2022)
The miniature CRISPR-Cas12m effector binds DNA to block transcription
Mol.Cell,
82
(23)
Author count: 6
Sabeti Azad M., Cardoso Batista A., Faulon J.L., Beisel C.L., Bonnet J., Kushwaha M.
(2022)
Cell-Free Protein Synthesis from Exonuclease-Deficient Cellular Extracts Utilizing Linear DNA Templates
J.Vis.Exp.
(186)
Author count: 6
Vialetto E., Yu Y., Collins S.P., Wandera K.G., Barquist L., Beisel C.L.
(2022)
A target expression threshold dictates invader defense and prevents autoimmunity by CRISPR-Cas13
Cell Host.Microbe
Author count: 2
Jiao C., Beisel C.L.
(2022)
Reprogramming TracrRNAs for Multiplexed RNA Detection
,
2518
(13)
Author count: 8
Wandera K.G., Alkhnbashi O.S., Bassett H.V.I., Mitrofanov A., Hauns S., Migur A., ... , Backofen R., Beisel C.L.
(2022)
Anti-CRISPR prediction using deep learning reveals an inhibitor of Cas13b nucleases
Mol.Cell
Author count: 9
Gallo M., Vento J.M., Joncour P., Quagliariello A., Maritan E., Silva-Soares N.F., Battistolli M., ... , Beisel C.L., Martino M.E.
(2022)
Beneficial commensal bacteria promote Drosophila growth by downregulating the expression of peptidoglycan recognition proteins
iScience.,
25
(6)
Author count: 2
Vento J.M., Beisel C.L.
(2022)
Genome Editing with Cas9 in Lactobacilli
Methods Mol.Biol.,
2479
(16)
Author count: 2
Sparmann A., Beisel C.L.
(2022)
CRISPR memories in single cells
Mol.Syst.Biol.,
18
(4)
Author count: 11
Liao C., Sharma S., Svensson S.L., Kibe A., Weinberg Z., Alkhnbashi O.S., Bischler T., Backofen R., Caliskan N., ... , Sharma C.M., Beisel C.L.
(2022)
Spacer prioritization in CRISPR -
Nat.Microbiol.,
7
(4)
Author count: 4
Wimmer F., Mougiakos I., Englert F., Beisel C.L.
(2022)
Rapid cell-free characterization of multi-subunit CRISPR effectors and transposons
Mol.Cell,
82
(6)
Author count: 3
Wimmer F., Englert F., Beisel C.L.
(2022)
A TXTL-Based Assay to Rapidly Identify PAMs for CRISPR-Cas Systems with Multi-Protein Effector Complexes
Methods Mol.Biol.
(24)
Author count: 12
Batista A.C., Levrier A., Soudier P., Voyvodic P.L., Achmedov T., Reif-Trauttmansdorff T., Devisch A., Cohen-Gonsaud M., Faulon J.L., Beisel C.L., ... , Bonnet J., Kushwaha M.
(2021)
Differentially Optimized Cell-Free Buffer Enables Robust Expression from Unprotected Linear DNA in Exonuclease-Deficient Extracts
ACS Synth.Biol.
Author count: 10
Gallo G., Mougiakos I., Bianco M., Carbonaro M., Carpentieri A., Illiano A., Pucci P., Bartolucci S., ... , van der Oost J., Fiorentino G.
(2021)
A Hyperthermoactive-Cas9 Editing Tool Reveals the Role of a Unique Arsenite Methyltransferase in the Arsenic Resistance System of Thermus thermophilus HB27
MBio.,
12
(6)
Author count: 2
Wandera K.G., Beisel C.L.
(2022)
Rapidly Characterizing CRISPR-Cas13 Nucleases Using Cell-Free Transcription-Translation Systems
Methods in Molecular Biology
(7)
Author count: 2
Gupta D., Beisel C.L.
(2021)
Illuminating the path to DNA repair
Cell,
184
(22)
Author count: 2
Liao C., Beisel C.L.
(2021)
The tracrRNA in CRISPR Biology and Technologies
Annu.Rev.Genet.
Author count: 6
Yu T., Zhang S., Matei R., Marx W., Beisel C.L., Wei Q.
(2021)
Coupling smartphone and CRISPR-Cas12a for digital and multiplexed nucleic acid detection
AIChE J.
Author count: 10
Taxiarchi C., Beaghton A., Don N.I., Kyrou K., Gribble M., Shittu D., Collins S.P., Beisel C.L., ... , Galizi R., Crisanti A.
(2021)
A genetically encoded anti-CRISPR protein constrains gene drive spread and prevents population suppression
Nature Communications,
12
(1)
Author count: 2
Mougiakos I., Beisel C.L.
(2021)
CRISPR transposons on the move
Cell Host and Microbe,
29
(5)
Author count: 12
Jiao C., Sharma S., Dugar G., Peeck N.L., Bischler T., Wimmer F., Yu Y., Barquist L., Schoen C., Kurzai O., ... , Sharma C.M., Beisel C.L.
(2021)
Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9
Science,
372
(6545)
Author count: 7
Durmusoglu D., Al'Abri I.S., Collins S.P., Cheng J., Eroglu A., ... , Beisel C.L., Crook N.
(2021)
In Situ Biomanufacturing of Small Molecules in the Mammalian Gut by Probiotic Saccharomyces boulardii
ACS Synth.Biol.,
10
(5)
Author count: 2
Luo M.L., Beisel C.
(2016)
SBE Supplement: Synthetic Biology - Engineering Genes with CRISPR-Cas9
AIChE Journal CEP Mag
(September 2016)
Author count: 4
Collins S.P., Rostain W., Liao Chunyu, Beisel C.L.
(2021)
Sequence-independent RNA sensing and DNA targeting by a split domain CRISPR-Cas12a gRNA switch
Nucleic Acids Res.,
49
(5)
Author count: 2
Alper H.S., Beisel C.L.
(2018)
Advances in CRISPR Technologies for Microbial Strain Engineering
Biotechnol.J.,
13
(9)
Author count: 1
Beisel C.L.
(2018)
CRISPR tool puts RNA on the record
Nature,
562
(7727)
Author count: 18
Annas G.J., Beisel C.L., Clement K., Crisanti A., Francis S., Galardini M., Galizi R., Grünewald J., Immobile G., Khalil A.S., Müller R., Pattanayak V., Petri K., Paul L., Pinello L., Simoni A., ... , Taxiarchi C., Joung J.K.
(2021)
A Code of Ethics for Gene Drive Research
CRISPR.J.,
4
(1)
Author count: 4
Miesler T., Wimschneider C., Brem A., Meinel L.
(2020)
Frugal Innovation for Point-of-Care Diagnostics Controlling Outbreaks and Epidemics
ACS Biomater.Sci.Eng,
6
(5)
Author count: 2
Collias D., Beisel C.L.
(2021)
CRISPR technologies and the search for the PAM-free nuclease
Nat.Commun.,
12
(1)
Author count: 3
Marshall R., Beisel C.L., Noireaux V.
(2020)
Rapid Testing of CRISPR Nucleases and Guide RNAs in an E.coli Cell-Free Transcription-Translation System
STAR.Protoc.,
1
(1)
Author count: 8
Collias D., Leenay R.T., Slotkowski R.A., Zuo Z., Collins S.P., McGirr B.A., ... , Liu J., Beisel C.L.
(2020)
A positive, growth-based PAM screen identifies noncanonical motifs recognized by the S. pyogenes Cas9
Sci.Adv.,
6
(29)
Author count: 2
Collins S.P., Beisel C.L.
(2020)
Your Base Editor Might Be Flirting with Single (Stranded) DNA: Faithful On-Target CRISPR Base Editing without Promiscuous Deamination
Mol.Cell,
79
(5)
Author count: 9
Orsi E., Mougiakos I., Post W., Beekwilder J., Dompe M., Eggink G., van der Oost J., ... , Kengen S.W.M., Weusthuis R.A.
(2020)
Growth-uncoupled isoprenoid synthesis in Rhodobacter sphaeroides
Biotechnol.Biofuels.,
13
(123)
Author count: 6
Jacobsen T., Ttofali F., Liao C., Manchalu S., Gray B.N., Beisel C.L.
(2020)
Characterization of Cas12a nucleases reveals diverse PAM profiles between closely-related orthologs
Nucleic Acids Res,
48
(10)
Author count: 6
Jacobsen T., Yi G., Al Asafen H., Jermusyk A.A., Beisel C.L., Reeves G.T.
(2020)
Tunable self-cleaving ribozymes for modulating gene expression in eukaryotic systems
PLoS ONE,
15
(4)
Author count: 2
Wimmer F., Beisel C.L.
(2020)
CRISPR-Cas Systems and the Paradox of Self-Targeting Spacers
Front.Microbiol.,
10
Author count: 5
Collias D., Marshall R., Collins S.P., Beisel C.L., Noireaux V.
(2019)
An educational module to explore CRISPR technologies with a cell-free transcription-translation system
Synth.Biol.,
4
(1)
Author count: 1
Beisel C.L.
(2020)
Methods for characterizing, applying, and teaching CRISPR-Cas systems
Methods,
172
Author count: 9
Siedler S., Rau M.H., Bidstrup S., Vento J.M., Aunsbjerg S.D., Bosma E.F., McNair L.M., ... , Beisel C.L., Neves A.R.
(2020)
Competitive exclusion is a major bioprotective mechanism of lactobacilli against fungal spoilage in fermented milk products
Appl.Environ.Microbiol.,
86
(7)
Author count: 3
Liao C., Slotkowski R.A., Beisel C.L.
(2019)
CRATES: A one-step assembly method for Class 2 CRISPR arrays
Methods in Enzymology
(24)
Author count: 14
Pickar-Oliver A., Black J.B., Lewis M.M., Mutchnick K.J., Klann T.S., Gilcrest K.A., Sitton M.J., Nelson C.E., Barrera A., Bartelt L.C., Reddy T.E., Beisel C.L., ... , Barrangou R., Gersbach C.A.
(2019)
Targeted transcriptional modulation with type I CRISPR-Cas systems in human cells
Nature Biotechnology,
37
(12)
Author count: 8
Liao C., Ttofali F., Slotkowski R.A., Denny S.R., Cecil T.D., Leenay R.T., ... , Keung A.J., Beisel C.L.
(2019)
Modular one-pot assembly of CRISPR arrays enables library generation and reveals factors influencing crRNA biogenesis
Nat.Commun.,
10
(1)
Author count: 6
Wandera K.G., Collins S.P., Wimmer F., Marshall R., Noireaux V., Beisel C.L.
(2020)
An enhanced assay to characterize anti-CRISPR proteins using a cell-free transcription-translation system
Methods,
172
Author count: 3
Vento J.M., Crook N., Beisel C.L.
(2019)
Barriers to genome editing with CRISPR in bacteria
J.Ind.Microbiol.Biotechnol.,
46
(9-10)
Author count: 3
Jacobsen T., Liao C., Beisel C.L.
(2019)
The Acidaminococcus sp. Cas12a nuclease recognizes GTTV and GCTV as non-canonical PAMs
FEMS Microbiol Lett.,
366
(8)
Author count: 3
Garenne D., Beisel C.L., Noireaux V.
(2019)
Characterization of the all-E. coli transcription-translation system myTXTL by mass spectrometry
Rapid Commun.Mass Spectrom.,
33
(11)
Author count: 6
Leenay R.T., Vento J.M., Shah M., Martino M.E., Leulier F., Beisel C.L.
(2019)
Genome Editing with CRISPR-Cas9 in Lactobacillus plantarum Revealed That Editing Outcomes Can Vary Across Strains and Between Methods
Biotechnol.J.,
14
(3)
Author count: 11
Westbrook A., Tang X., Marshall R., Maxwell C.S., Chappell J., Agrawal D.K., Dunlop M.J., Noireaux V., Beisel C.L., ... , Lucks J., Franco E.
(2019)
Distinct timescales of RNA regulators enable the construction of a genetic pulse generator
Biotechnol.Bioeng.,
116
(5)
Author count: 4
Liao C., Slotkowski R.A., Achmedov T., Beisel C.L.
(2019)
The Francisella novicida Cas 12a is sensitive to the structure downstream of the terminal repeat in CRISPR arrays
RNA Biology,
16
(4)
Author count: 9
Martino M.E., Joncour P., Leenay R., Gervais H., Shah M., Hughes S., Gillet B., ... , Beisel C., Leulier F.
(2018)
Bacterial Adaptation to the Host's Diet Is a Key Evolutionary Force Shaping Drosophila-Lactobacillus Symbiosis
Cell Host and Microbe,
24
(1)
Author count: 10
Agrawal D.K., Tang X., Westbrook A., Marshall R., Maxwell C.S., Lucks J., Noireaux V., Beisel C.L., ... , Dunlop M.J., Franco E.
(2018)
Mathematical Modeling of RNA-Based Architectures for Closed Loop Control of Gene Expression
ACS Synth.Biol.,
7
(5)
Author count: 7
Dugar G., Leenay R.T., Eisenbart S.K., Bischler T., Aul B.U., ... , Beisel C.L., Sharma C.M.
(2018)
CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9
Mol.Cell,
69
(5)
Author count: 5
Maxwell C.S., Jacobsen T., Marshall R., Noireaux V., Beisel C.L.
(2018)
A detailed cell-free transcription-translation-based assay to decipher CRISPR protospacer-adjacent motifs
Methods,
143