Our Research
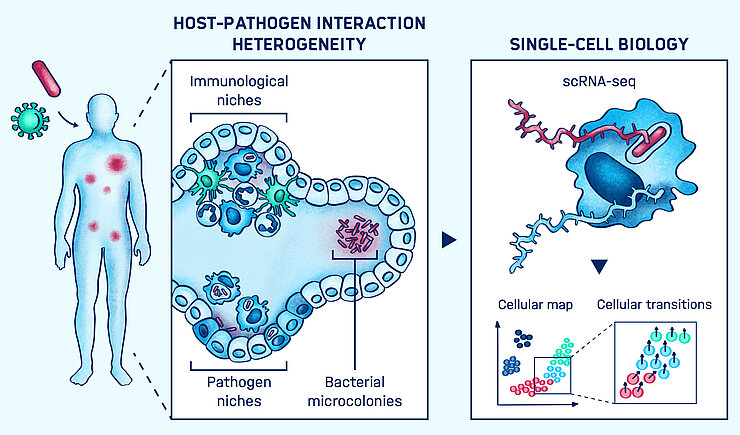
Persistent bacterial infections are caused by a minor subpopulation of intracellular pathogens, called ‘persisters’, that reside in different cell types and tissue locations for years asymptomatically. These subpopulations establish specific cellular organization to enable them to evade immune surveillance and chemotherapy treatments. Histological studies have described complex tissue remodeling during the infection and emerging in vivo studies at the single-cell level have begun to reveal the heterogeneity of infection foci.
However, the cellular architecture of the infection foci and which are the favorite niches among this complex anatomical tissue landscape that impact disease outcome remain open questions. For example, Salmonella spp. are believed to reside in a large variety of cells including macrophages, neutrophils, dendritic cells and epithelial cells. These large cell types exist as a myriad of different sub-classes, which were, until recently, not appreciated. Similarly, within an infected tissue such as the spleen, many infected cells escape inflammatory lesions and disseminate into tissues. Therefore, single-cell studies in an in vivo context are necessary to understand the heterogeneity inherent in infected cells, their microenvironment and their function. The Single-Cell Analysis group develop and combine in vitro and in vivo single-cell transcriptomics to decipher the cellular organization of infection foci and their functional consequences on infection outcome.
The recent emergence of single-cell genome-wide transcriptomics is proving to be a powerful approach to decipher both cellular identities and function making possible to study heterogeneity. It is being facilitated by the development of automated platforms that enable the processing of hundreds and thousands of single-cells in parallel. In the context of infection, we have pioneered the use of single-cell RNA-seq to investigate heterogeneity in the response of mouse bone-marrow-derived macrophages to Salmonella, focusing on bacteria with different growth status including non-growing ‘persisters’ that have been linked to recurrent infections. We have described how Salmonella impact the wide spectrum of host polarization and revealed the existence of a subset of macrophages that escape inflammatory and immune activation programs. While providing new insights into the host response, the study was limited to analyzing infected cells from in vitro mono-cultures. The next step is now to decipher the response of single cells of infected tissues, which remains an unmet challenge.
Moreover, the Single-cell Analysis group is fully committed to developing the full potential of single-cell RNA-seq for addressing fundamental scientific questions of infection biology in general, at the HIRI as well as other locations of the HZI.
Our Research
Persistent bacterial infections are caused by a minor subpopulation of intracellular pathogens, called ‘persisters’, that reside in different cell types and tissue locations for years asymptomatically. These subpopulations establish specific cellular organization to enable them to evade immune surveillance and chemotherapy treatments. Histological studies have described complex tissue remodeling during the infection and emerging in vivo studies at the single-cell level have begun to reveal the heterogeneity of infection foci.
However, the cellular architecture of the infection foci and which are the favorite niches among this complex anatomical tissue landscape that impact disease outcome remain open questions. For example, Salmonella spp. are believed to reside in a large variety of cells including macrophages, neutrophils, dendritic cells and epithelial cells. These large cell types exist as a myriad of different sub-classes, which were, until recently, not appreciated. Similarly, within an infected tissue such as the spleen, many infected cells escape inflammatory lesions and disseminate into tissues. Therefore, single-cell studies in an in vivo context are necessary to understand the heterogeneity inherent in infected cells, their microenvironment and their function. The Single-Cell Analysis group develop and combine in vitro and in vivo single-cell transcriptomics to decipher the cellular organization of infection foci and their functional consequences on infection outcome.
The recent emergence of single-cell genome-wide transcriptomics is proving to be a powerful approach to decipher both cellular identities and function making possible to study heterogeneity. It is being facilitated by the development of automated platforms that enable the processing of hundreds and thousands of single-cells in parallel. In the context of infection, we have pioneered the use of single-cell RNA-seq to investigate heterogeneity in the response of mouse bone-marrow-derived macrophages to Salmonella, focusing on bacteria with different growth status including non-growing ‘persisters’ that have been linked to recurrent infections. We have described how Salmonella impact the wide spectrum of host polarization and revealed the existence of a subset of macrophages that escape inflammatory and immune activation programs. While providing new insights into the host response, the study was limited to analyzing infected cells from in vitro mono-cultures. The next step is now to decipher the response of single cells of infected tissues, which remains an unmet challenge.
Moreover, the Single-cell Analysis group is fully committed to developing the full potential of single-cell RNA-seq for addressing fundamental scientific questions of infection biology in general, at the HIRI as well as other locations of the HZI.
Dr Antoine-Emmanuel Saliba
Single-cell sequencing is a terrific scientific revolution equivalent to genome sequencing. In ten years from now, all biology including infection biology will turn to be quantitative, predictive and mostly based on single-cell.

Antoine-Emmanuel Saliba performed his undergraduate studies in biochemical engineering at the INSA Toulouse (France) and performed his PhD work under the supervision of Jean-Louis Viovy at the Institut Curie (Paris, France). During this time he pioneered the development of microfluidics-based integrated systems to sort and analyse rare cancer cell subpopulations. As postdoctoral EIPOD-fellow within the European Molecular Biology Laboratory (EMBL, Heidelberg, Germany), he developed one of the first systems-biology approaches to track protein-lipid interactions on the genome-scale. After this period, he joined the Vogel lab in Würzburg as senior postdoc where he introduced the use of single-cell RNA-seq to track the fate of individual infected cells with Salmonella enterica. Since 2017, he leads the Single-cell Analysis group within HIRI.
Selected Publications
Cochain C*, Vafadarnejad E*, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE*, Zernecke A*
Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis
Circulation Res 2018, DOI: 10.1161/CIRCRESAHA.117.312509
Saliba AE, C Santos S, Vogel J
New RNA-seq approaches for the study of bacterial pathogens
Curr Opin Microbiol 2017, 35:78-87
Saliba AE, Li L, Westermann AJ, Appenzeller S, Stapels DA, Schulte LN, Helaine S, Vogel J
Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella
Nature Microbiol 2016, 2:16206
Saliba AE, Vonkova I, Deghou S, Ceschia S, Tischer C, Kugler KG, Bork P, Ellenberg J, Gavin AC
A protocol for the systematic and quantitative measurement of protein-lipid interactions using the liposome-microarray-based assay
Nature Protoc 2016, 11(6): 1021-1038
Vonkova I*, Saliba AE*, Deghou S, Anand K, Ceschia S, Doerks T, Galih A, Kugler KG, Maeda K, Rybin V, van Noort V, Ellenberg J, Bork P, Gavin AC
Lipid Cooperativity as a General Membrane-Recruitment Principle for PH Domains
Cell Rep 2015, 12(9): 1519-1530